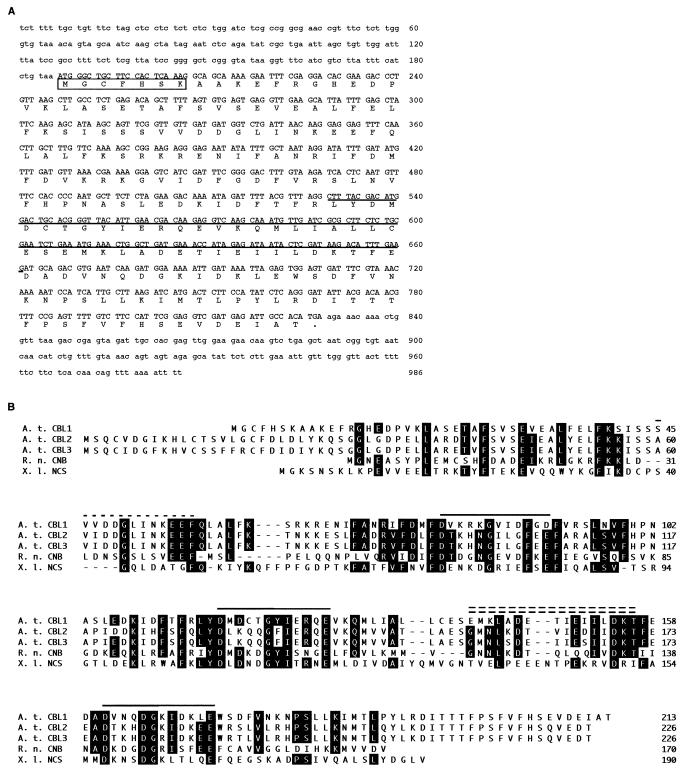
Figure 1.
Sequence analyses of AtCBL1 cDNA and AtCBL proteins. (A) cDNA and deduced amino acid sequences of AtCBL1. The coding region of the cDNA is presented in capital letters and the noncoding regions in lowercase letters. The boxed sequence contains a typical myristoylation site (MGXXXSK). The underlined sequence represents the cDNA fragment identified by PCR (see Materials and Methods). An in-frame stop codon (taa) is immediately upstream from the ATG starting codon. (B) Alignment of the amino acid sequences of three AtCBL proteins from Arabidopsis (A. t. CBL1, A. t. CBL2, A. t. CBL3) with rat CNB (R. n. CNB), and Xenopus neuronal calcium sensor (X. l. NCS). Alignment was generated by using the clustal method with dnastar software. Amino acids identical to the consensus sequence of the alignment are shown on black background. The numbers on the right indicate the amino acid position. Dashes indicate gaps introduced to improve the alignment. Solid lines above the sequence indicate position of EF-hand regions. The single-dashed line represents a variation of the Ca2+-binding domain. The double-dashed line denotes the CNA interaction domain.