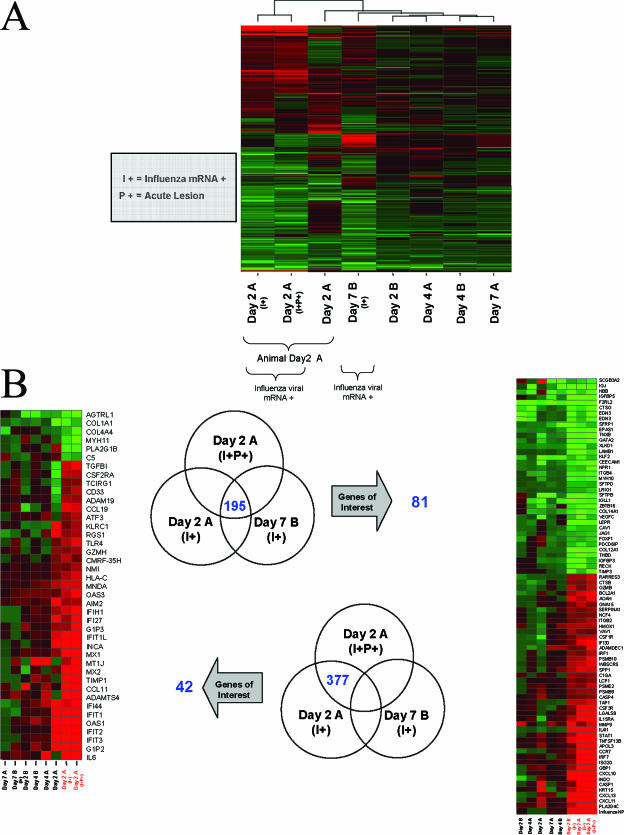
FIG. 2.
(A) Unsupervised clustering of gene expression profiles. The profiles for animal Day 2 A show similarities based on the fact that they were sampled from the same animal, while viral-mRNA-positive samples showed similarities as well. Day 2 A (I+P+) was positive for viral mRNA and within the main lesion, Day 2 A (I+) was positive for viral mRNA and adjacent to the lesion, and Day 2 A was negative for viral mRNA and adjacent to the lesion as well. All gene expression profiles are the results of comparing gene expression in the lungs of individual experimental animals versus gene expression in the lungs of mock-infected animals (pooled), and genes were included if they met the criterion of a twofold or greater change (P ≤ 0.01). A two-of-eight strategy allowed samples to cluster together if profile similarities existed based on timing of inoculation (n = 2 samples for each day). A three-of-eight strategy allowed samples to cluster together if profile similarities existed based on the presence or absence of viral mRNA (n = 3 samples with viral mRNA) or based on the samples being from the same animal (n = 3 samples from animal Day 2 A). Very similar clusters were obtained in both cases, so only one is represented here. (B) Venn diagrams showing genes with changes of ≥2-fold (P ≤ 0.01) in samples Day 2 A (I+P+), Day 2 A (I+), and Day 7 B (I+) (upper Venn diagram) and in samples Day 2 A (I+P+) and Day 2 A (I+) but not in sample Day 7 B (I+) (lower Venn diagram). Although there were fewer genes within the intersection of the three samples (195 genes), a greater proportion of these genes was annotated and had functions relevant to the immune response, infectious disease, or pulmonary stress. Heat maps show the same genes regulated in day 2, 4, and 7 lung samples. All gene expression profiles are the results of comparing gene expression in the lungs of individual experimental animals versus gene expression in the lungs of mock-infected animals (pooled).