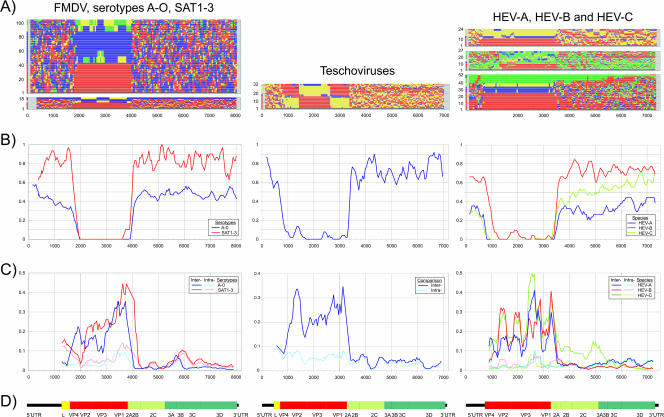
FIG. 1.
(A) Ordering (y axis) of variants assigned to different serotypes in phylogenetic trees generated from consecutive 300-base fragments across complete genome sequence alignments of aphthoviruses (left), teschovirus (middle), and human enteroviruses (right). Serotypes are color labeled as follows. For serotypes A to O of FMDV, serotype A is red, B is yellow, C is green, and O is blue; for SAT serotypes, SAT-1 is red, SAT-2 is yellow, and SAT-3 is blue; and for teschoviruses, serogroup 1 is red (serotypes 1, 3, 10, and 11), serogroup 2 is yellow (serotypes 9, 7, and 5), and serogroup 3 is blue (serotypes 6, 2, 4, and 8). For human enteroviruses, the following serotypes were labeled. For HEV-A, enterovirus 71 is red, CAV-16 is blue, and others are yellow; for HEV-B, echovirus 11 is red, echovirus 9 is blue, CBV-5 is yellow, and others are green; for HEV-C, PV1 is red, PV2 is blue, PV3 is green, and others are blue. (B) Segregation scores for consecutive fragments across genomes, where 0 (y axis) represents perfect phylogenetic segregation by assigned group (serotypes) and 1 represents the absence of association between phylogeny and group assignment. Similar results for slightly different sequence subsets of enteroviruses HEV-A to HEV-C have been presented previously (55) and are included for comparison. (C) Mean pair-wise amino acid sequence distances between sequences in consecutive 300-base fragments across the genome. Values are averaged over a window size of 3. Separate mean values were calculated for sequence comparisons within and between groups (pale and dark colors). (D) Genome diagrams of FMDV, teschoviruses, and human enteroviruses drawn to scale and numbered according to the reference sequences NC_011450 for FMDV, PEN011380 for teschoviruses, and POL3L37 for enteroviruses.