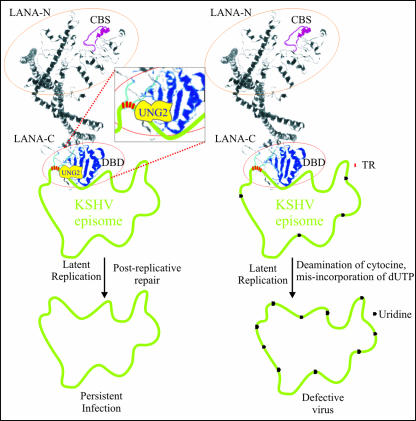
FIG. 9.
Proposed model of LANA-UNG2 interaction at KSHV TRs. LANA is a large (1,162-amino-acid) nuclear protein important for tethering of the viral genome to the host chromosome (4, 16). The three-dimensional structure of LANA, predicted with Robetta, a free protein structure prediction server (http://robetta.bakerlab.org/), shows distinct amino- and carboxyl-terminal domains. The amino-terminal domain of LANA shows a distinct chromosome binding sequence (CBS, amino acids 5 to 22). The DNA binding domain (DBD) of LANA, which lies between amino acids 996 and 1139 (38), is marked in blue (LANA-C). The interaction of LANA with UNG2 was mapped to the carboxyl terminus (amino acids 945 to 1162). The interaction of LANA and UNG2 is most likely important for bringing UNG2 into close proximity to the TR DNA, which is most likely important for the removal of any uracil residues incorporated during the replication process. Incorporation of uracil, if left unrepaired, may lead to the production of defective virus. PEL cells depleted of UNG2 with shRNA showed reduced KSHV genomic copies, possibly because of the accumulation of mutations caused by unrepaired incorporation of uracil residues during successive rounds of replication.