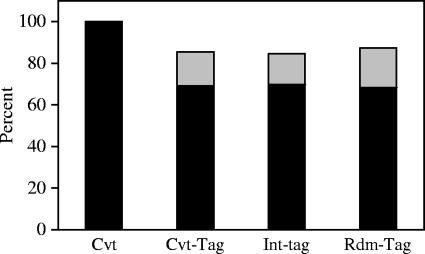
FIG. 5.
Mapping of short integration sequences to unique locations and those in identifiable repeat elements. Integration site sequences from cells infected with NL-Mme were determined using the conventional assay (Cvt) or the high-throughput assay (Int-tag). Each sequence was mapped to either unique locations (black bars) or identifiable repeat elements (gray bars), and the results are expressed as percentages of total integration site sequences. The results were compared with those obtained from conventional tags (Cvt-tag), generated by taking the first 19 or 20 nucleotides immediately adjacent to the viral DNA of each integration site sequence determined by the conventional assay, or a random library of 19- and 20-bp cellular sequences (Rdm-tag) generated in silico.