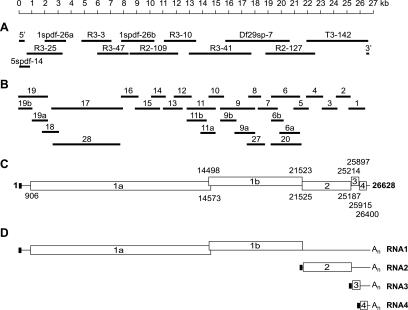
FIG. 2.
Structural organization and sequence analysis of the WBV (strain DF24/00) genome. (A) Given are the sizes and positions of cDNA clones from WBV genomic libraries that were used to determine the WBV genome sequence. Also shown are the 5′- and 3′-terminal amplicons generated by RACE. (B) Given are the sizes and positions of RT-PCR products used to ascertain the sequence derived from the cDNA clones shown in panel A. (C) Predicted functional ORFs in the WBV genome. Numbers indicate the 5′ and 3′ nucleotides, respectively, of predicted translation start and stop codons. Note that translation of ORF1b is predicted to involve a −1 ribosomal frameshift occurring just upstream of the ORF1a translation stop codon (and downstream of the most 5′-terminal AUG codon that is used here to indicate the ORF1b 5′ end) (for further details, see the text and Table 1). (D) WBV-specific RNAs as determined in this study. The little black box at the 5′ end of the genome indicates the 42-nt leader sequence, which is also present at the 5′ ends of the three subgenome-length RNAs (Fig. 3 and 4). The available evidence from other nidoviruses (37, 47) suggests that attachment of the leader sequence to the coding (body) sequences of WBV subgenome-length RNAs is due to discontinuous extension of subgenome-length minus-strand RNAs. In this process, nascent minus strands switch their template at TRSs located upstream of the S, M, and N genes and bind then to an identical sequence called leader TRS near the 5′ end of the genome, after which the leader sequence is copied to complete negative-strand synthesis (see the text and Fig. 4).