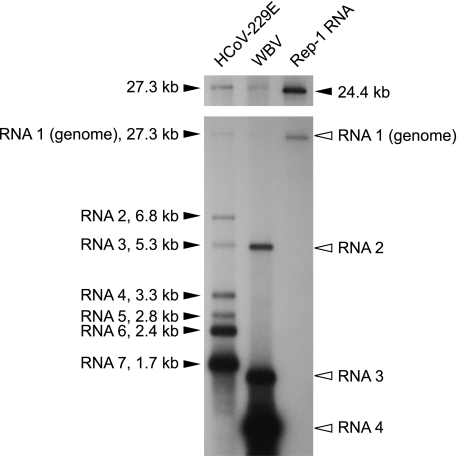
FIG. 3.
Detection of WBV genome- and subgenome-length RNAs in virus-infected cells. Northern blot analysis of poly(A)-containing RNA isolated from WBV-infected EPC cells (lane 2). Poly(A) RNAs isolated from HCoV-229E-infected MRC-5 cells (lane 1) and HCoV-229E-derived replicon RNA Rep-1 (lane 3) (28) were used as RNA size markers in this experiment. To detect both the HCoV-229E- and WBV-specific RNAs, a mixture of α-32P-multiprime-labeled probes specific for the 3′-terminal regions of HCoV-229E (nucleotides 26857 to 27277) and WBV (nucleotides 25992 to 26582) was used for hybridization. HCoV-229E genome- and subgenome-length RNAs and the in vitro-transcribed HCoV-229E Rep-1 RNA are indicated by black arrowheads, with sizes given in kilobases. White arrowheads indicate the four WBV-specific RNAs detected in this experiment. The longer exposure presented above shows the size of the WBV genomic RNA more clearly and allows its size to be compared with those of the 27.3- and 24.4-kb marker RNAs. The calculated sizes (Table 1 and Fig. 4) of the sg RNAs are 5,162 nts (RNA 2), 1,475 nts (RNA 3), and 774 nts (RNA 4) [including the 5′ leader but excluding the poly(A) tail].