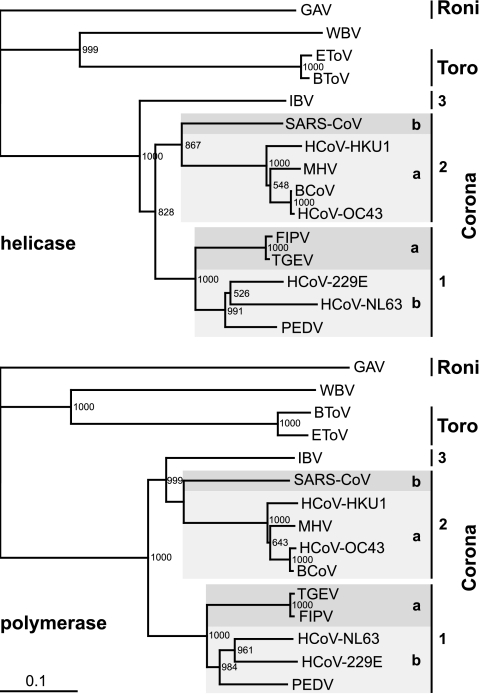
FIG. 8.
Phylogenetic analysis of WBV helicase and polymerase core domains. Phylogenetic trees were generated from multiple-sequence alignments of the most conserved regions of nidovirus RNA-dependent RNA polymerase (residues Thr4723 to Gln5396 in the WBV pp1ab sequence) and helicase domains (residues Ala5644 to Cys5924 in the WBV pp1ab sequence), using the neighbor-joining algorithm as implemented in the ClustalX 1.8 program (for details, see Materials and Methods). The WBV sequences were compared with those from Gill-associated virus (GAV, accession no. AF227196), Equine torovirus Berne (EToV, X52374), and Bovine torovirus Breda-1 (BToV, AY427798) as well as from viruses representing the three coronavirus groups, including the recently introduced subgroups 1a, 1b, 2a, and 2b (26). Group 1a, Transmissible gastroenteritis virus Purdue-115 (TGEV, accession no. Z34093) and Feline infectious peritonitis virus WSU 79/1146 (FIPV, DQ010921); group 1b, HCoV-229E (NC_002645), Human coronavirus NL63 Amsterdam I (HCoV-NL63, AY567487), and Porcine epidemic diarrhea virus CV777 (PEDV, AF353511); group 2a, Bovine coronavirus LUN (BCoV, AF391542), Human coronavirus OC43 serotype Paris (HCoV-OC43, AY585229), Mouse hepatitis virus A59 (MHV, NC_001846), and Human coronavirus HKU1 (HCoV-HKU1, NC_006577); group 2b, Severe acute respiratory syndrome coronavirus Frankfurt 1 (SARS-CoV, AY291315); group 3, Avian infectious bronchitis virus Beaudette (IBV, NC_001451).