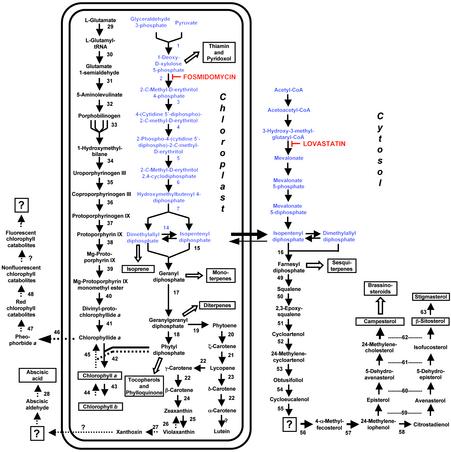
Fig. 1.
Overview of isoprenoid metabolic pathways localized to the cytosol and to plastids in plants, with an emphasis on the metabolism of chlorophylls, carotenoids, and sterols. The symbol for anabolic reactions is a solid arrow, and catabolic reactions are indicated by arrows with dotted lines. Open arrows depict multiple enzymatic steps, and key metabolites are boxed. Known enzymes involved in isoprenoid metabolism are numbered, and question marks indicate steps for which an enzymatic activity has not yet been demonstrated. Evidence for the branching of the plastidial MVA-independent pathway to yield IPP and dimethylallyl diphosphate independently has recently been reported (27). The following enzymes are represented: 1, 1-deoxy-d-xylulose 5-phosphate synthase; 2, DXR; 3, 2-C-methyl-d-erythritol 4-phosphate cytidyltransferase; 4, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; 5, 2,4-C-methyl-d-erythritol cyclodiphosphate synthase; 6, 1-hydroxy-2-methyl-2-(E)-butenyl-4-phosphate synthase; 7, 1-hydroxy-2-methyl-2-(E)-butenyl-4-phosphate reductase; 8, acetoacetyl-CoA thiolase; 9, 3-hydroxy-3-methylglutaryl-CoA synthase; 10, HMGR; 11, MVA kinase; 12, phospho-MVA kinase; 13, MVA diphosphate decarboxylase; 14, IPP: dimethylallyl diphosphate isomerase; 15, geranyl diphosphate synthase; 16, farnesyl diphosphate synthase; 17, geranylgeranyl diphosphate synthase; 18, geranylgeranyl reductase; 19, phytoene synthase; 20, phytoene desaturase; 21, ζ-carotene desaturase; 22, lycopene β-cyclase; 23, lycopene ε-cyclase; 24, β-carotene hydroxylase; 25, zeaxanthin epoxidase; 26, violaxanthin de-epoxidase; 27, epoxycarotenoid (neoxanthin) cleavage enzyme; 28, abscisic aldehyde oxidase; 29, glutamyl tRNA synthetase; 30, glutamyl tRNA reductase; 31, glutamate 1-semialdehyde aminotransferase; 32, aminolevulinate dehydratase; 33, porphobilinogen deaminase; 34, uroporphyrinogen synthase; 35, uroporphyrinogen decarboxylase; 36, coproporphyrinogen III oxidase; 37, protoporphyrinogen IX oxidase; 38, Mg-protoporphyrinogen IX chelatase; 39, Mg-protoporphyrinogen IX methyltransferase; 40, protoporphyrinogen cyclase (enzyme activity not yet characterized in plants); 41, protochlorophyllide reductase; 42, chlorophyll synthetase; 43, chlorophyll a oxygenase; 44, chlorophyll b reductase (gene not cloned yet); 45, chlorophyllase; 46, de-chelatase (gene not cloned yet); 47, pheophorbide a oxygenase (gene not yet cloned); 48, red chlorophyll catabolite reductase; 49, squalene synthase; 50, squalene monooxygenase; 51, cycloartenol synthase; 52, cycloartenol C24 methyltransferase; 53, 24-methylenecycloartenol C-4 methyl oxidase; 54, cycloeucalenol cycloisomerase; 55, sterol C14 reductase; 56, obtusifoliol 14-demethylase; 57, C-8,7 sterol isomerase; 58, sterol C-methyltransferase 2; 59, sterol C-4 methyl oxidase; 60, sterol C5-desaturase; 61, sterol Δ7 reductase; 62, sterol C24-reductase; and 63, sterol C22-desaturase.