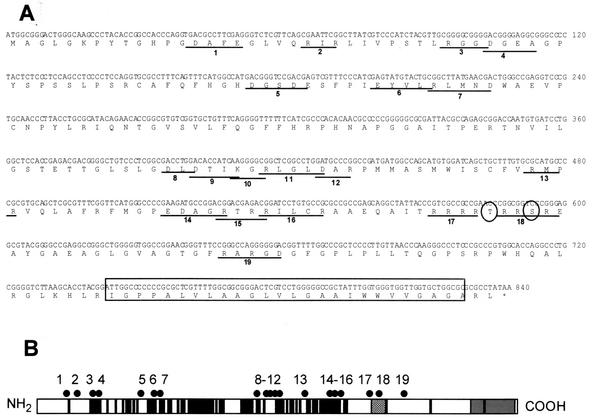
FIG. 1.
Locations of charged cluster mutations within the UL34 gene. (A) Nucleotide and amino acid sequences of the UL34 gene, showing the positions of charged cluster mutations. Each charged amino acid in a charged cluster was changed to an alanine. The location of a phosphorylation site recognized by US3 protein kinase is denoted by circles, and the putative transmembrane domain is indicated by the boxed region. (B) Schematic depiction of the sequence arrangement and location of charged clusters in the UL34 protein of HSV-1. The positions of the putative transmembrane domain (grey box), US3 phosphorylation consensus (hatched box), and residues invariant between HSV-1, varicella-zoster virus, and equine herpesvirus type 1 (black boxes) are indicated. Each of the black dots above the schematic represents the location of a charged cluster mutation within the gene. The mutations were numbered from 1 to 19, starting at the N terminus.