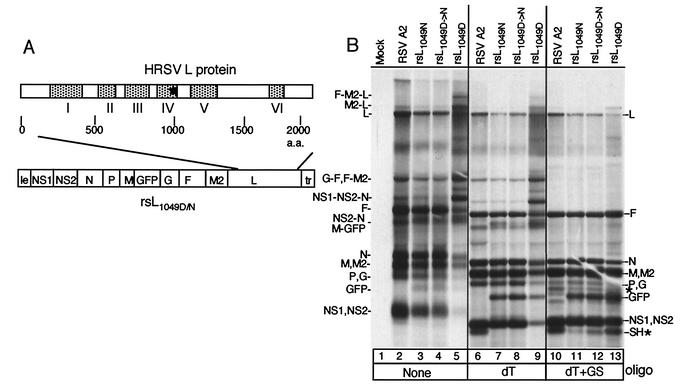
FIG. 1.
RNA synthesis of HRSV A2 and recovered engineered viruses. (A) Schematic of the rsL1049D or rsL1049N cDNA in which the SH coding region was replaced with that of GFP. To create rsL1049N, an SphI fragment in the L gene was replaced. Schematic of L protein illustrating the positions of the conserved domains is shown above the cDNA. An asterisk shows the location of amino acid 1049. Viruses were recovered by transfecting appropriate cDNAs along with the HRSV support plasmids pN, pP, pM2ORFI, and L1049D or L1049N into HEp-2 cells infected with MVA-T7. (B) HEp-2 cells were infected with the A2 strain of HRSV or individual engineered viruses, and viral RNAs were labeled 16 h postinfection with [3H]uridine in the presence of actinomycin D. Cells were infected at a multiplicity of infection of approximately 0.5 for each virus, except the rsL1049D virus, for which a multiplicity of infection of approximately 3 was used. RNAs were either undigested (lanes 1 to 5), digested with RNase H and 1 μg of oligo(dT) (lanes 6 to 9), or digested with RNase H, 1 μg of oligo(dT), and 1 μg of gene start oligonucleotide (lanes 10 to 13). Use of the gene start oligonucleotide resulted in some nonspecific cleavage, indicated by new bands (*) not present in the undigested samples or samples digested with oligo(dT) alone. The positions of monocistronic and polycistronic mRNAs are shown to the right and left side of the gel. Those separated by commas are monocistronic or polycistronic RNAs that comigrate. Those separated by hyphens denote polycistronic RNAs. dT, oligo(dT); GS, gene start oligonucleotide.