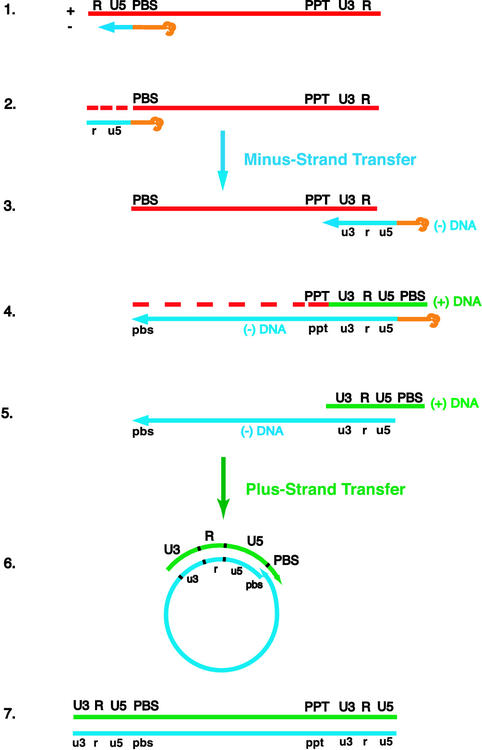
FIG. 1.
Schematic diagram of the events in reverse transcription. (Step 1) HIV reverse transcription is initiated by a cellular tRNA primer, tRNA3Lys, following annealing of the 3′ 18 nt of the tRNA to the 18-nt primer binding site (PBS) near the 5′ end of the genome. RT catalyzes synthesis of (−) SSDNA, which contains copies of the repeat (R) sequence and the unique 5′ genomic sequence (U5). (Step 2) As the primer is extended, the RNase H activity of RT degrades the genomic RNA sequences that have been reverse transcribed. (Step 3) (−) SSDNA is transferred to the 3′ end of viral RNA (minus-strand transfer). (Step 4) Elongation of minus-strand DNA and RNase H degradation continue. Plus-strand synthesis is initiated by the 15-nt PPT immediately upstream of the unique 3′ genomic sequence (U3). RT copies the u3, u5, and r regions in minus-strand DNA, as well as the 3′ 18 nt of the tRNA primer, thereby reconstituting the PBS. The product formed is termed (+) SSDNA. (Step 5) RNase H removal of the PPT and tRNA primers. (Step 6) Transfer of (+) SSDNA to the 3′ end of minus-strand DNA (plus-strand transfer) is followed by circularization of the two DNA strands and displacement synthesis. (Step 7) Minus- and plus-strand DNAs are elongated, resulting in a linear double-stranded DNA with a long terminal repeat at each end. Viral RNA is shown in red; minus-and plus-strand DNAs are shown in blue and green, respectively; and the tRNA primer is in orange. Minus-strand and plus-strand sequences are depicted in lower and upper case, respectively. The dashed red lines represent RNase H cleavage of genomic RNA.