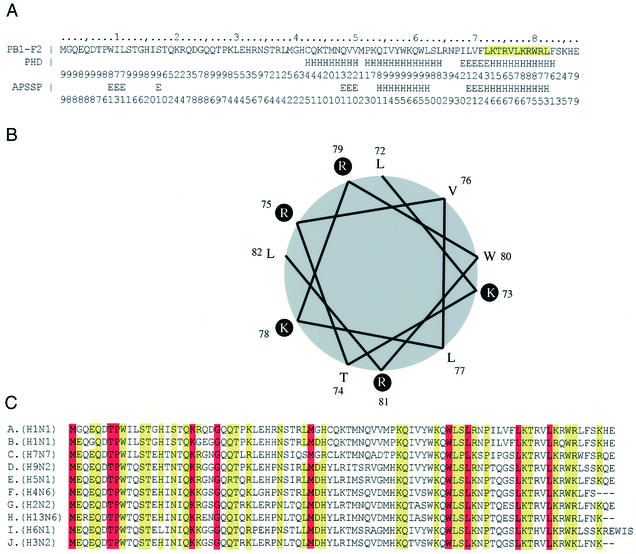
FIG. 1.
PB1-PB1-F2 sequence comparison and structural predictions. (A) The sequence of PB1-F2 is shown along with structural predictions (H, helix; E, extended) by PHD and APSSP programs with respect to helix formation. (B) Helical wheel representation of a predicted amphipathic helix formed by residues 72 to 82 of PB1-F2. (C) Sequences of PB1-F2 representing all IAV subtypes for which sequences are available. Residues present in all 90 IAV strains examined are highlighted in red; those conserved in >90% of the strains are highlighted in yellow. Strains are indicated as follows: A, Puerto Rico/8/34 (H1N1); B, FM/1/47 (H1N1); C, Equine/London/1416/73 (H7N7); D, Chicken/Korea/38349-p96323/96 (H9N2); E, Goose/Guangdong/1/96 (H5N1); F, Duck/Nanchang/662/98 (H4N6); G, Leningrad/134/47/57 (H2N2); H, Gull/Maryland/704/77 (H13N6); I, Teal/Hong Kong/W312/97 (H6N1); J, NT/60/68 (H3N2).