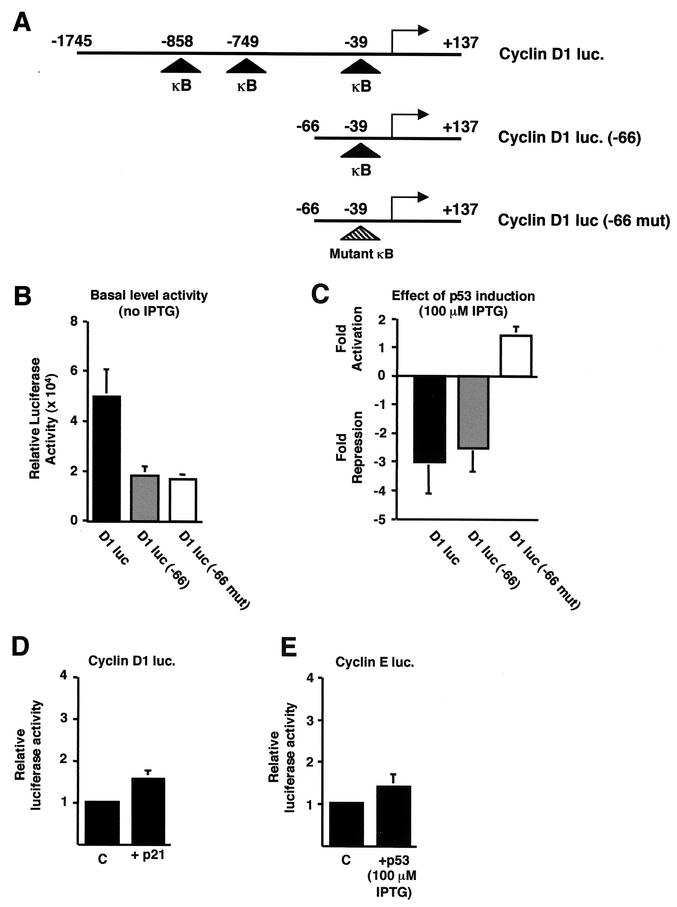
FIG.3.
p53 specifically inhibits cyclin D1 promoter activity. (A) Schematic diagram of the cyclin D1, cyclin D1 (−66), and cyclin D1 (−66 mut) luciferase reporter plasmids used. The relative positions of the previously described NF-κB binding sites are shown. luc, luciferase. (B) Relative luciferase activity of each of the cyclin D1-luciferase constructs. A quantity of 1.5 μg of each of the cyclin D1-luciferase reporter plasmids was transfected into H1299w/tp53 cells. (C) Inhibition of cyclin D1 promoter activity by p53 requires the proximal κB site. A quantity of 1.5 μg of each of the cyclin D1-luciferase reporter plasmids was transfected into H1299w/tp53 cells. p53 was induced by treatment with 100 μM IPTG, and cells were harvested 16 h after induction. Results are expressed as change in activation or repression (n-fold) relative to levels seen in the relevant untreated cell controls. (D) p21 does not inhibit cyclin D1 promoter activity in H1299w/tp53 cells. One and a half micrograms of cyclin D1-luciferase reporter plasmid was cotransfected with 1.5 μg of RSV-p21 or control RSV plasmid. Results are expressed as relative luciferase activity. (E) p53 does not inhibit cyclin E promoter activity in H1299w/tp53 cells. One and a half micrograms of cyclin E luciferase plasmid was transfected into H1299w/tp53 cells, and p53 expression was induced by 100 μM IPTG. Cells were harvested 16 h after IPTG treatment, and results are expressed as relative luciferase activity.