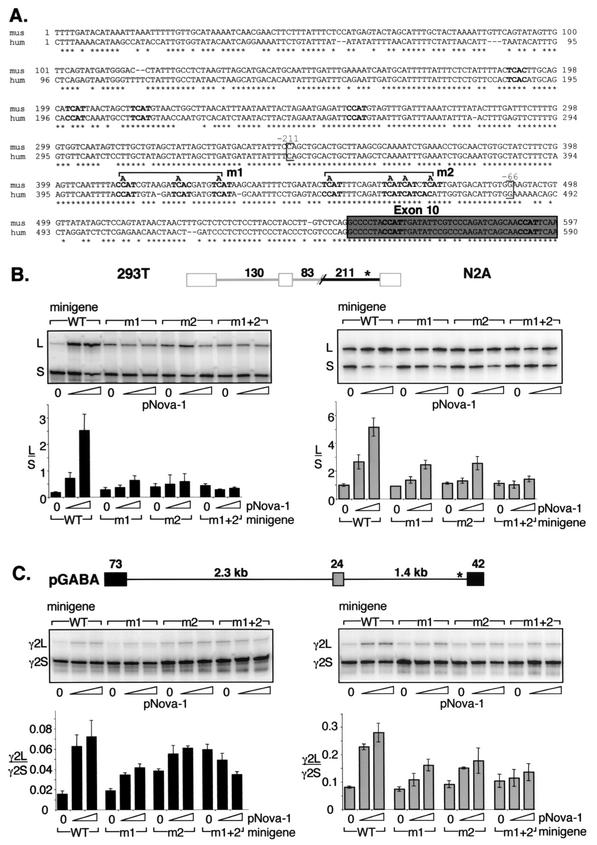
FIG.5.
Mutagenesis of the YCAY repeats close to E10 abolishes Nova-dependent regulation of GABAARγ2 minigene alternative splicing. (A) Genomic sequences upstream of GABAARγ2 exon 10 (gray box) from mouse (mus) and human (hum) were aligned by ClustalW alignment (MacVector). Conserved YCAY elements are shown in boldface characters. Brackets indicate the region corresponding to the in vitro transcribed RNA used as described for Fig. 6B and are numbered relative to exon 10. m1 and m2 represent mutations of three and four YCAY repeats to YAAY, respectively. (B) Chimeric minigenes containing the mutations depicted in panel A were cotransfected into 293T or N2A cells with increasing amounts of pNova-1 (0, 0.5, or 2.0 μg), and spliced products were measured by RT-PCR and phosphorimage analysis as described for Fig. 2. The transfected pNova-1 expression was monitored by Western blotting (data not shown). Quantitation of the data is shown below representative autoradiographs and is presented as L/S exon use ratios ± standard deviations for three independent transfections. WT, wild type. (C) GABAARγ2 minigenes containing the mutations depicted in panel A were cotransfected into 293T or N2A cells with increasing amounts of pNova-1 (0, 0.5, or 2.0 μg), and spliced products were measured by RT-PCR and phosphorimage analysis as described for Fig. 2. The transfected pNova-1 expression was monitored by Western blotting (data not shown). Quantitation of the data is shown below representative autoradiographs and is presented as γ2L/γ2S exon use ratios ± standard deviations for three independent transfections. WT, wild type.