TABLE 1.
Crystallographic data and refinement statistics
| Parameter | Value |
|---|---|
| PPAT:CoA data collection | |
| Resolution [Å] | 1.78 |
| Total data | 815,367 |
| Unique data | 39,181 |
| Overall completeness | 0.99 |
| Completeness (1.84-1.78 Å) | 0.907 |
| F2 > 3σ(F2) [%] | 0.76 |
| Rmergea (overall) | 0.036 |
| Rmerge (1.84-1.78 Å) | 0.257 |
| Avg F2/σ(F2) | 41.5 |
| Crystallographic refinement | |
| No. of reflections | 39,134 |
| Final model parameters | |
| No. of amino acid residues | 315 |
| No. of protein atoms | 2,481 |
| No. of solvent molecules | 286 |
| No. of substrate (sulfate) molecules | 2 (4) |
| Resolution range [Å] | 20-1.78 |
| R factorb overall | 0.218 |
| R factor (1.89-1.78 Å) | 0.302 |
| Rfreec overall | 0.241 |
| Rfree (1.89-1.78 Å) | 0.323 |
| Avg main chain B factor [Å2] | 21.6 |
| Avg side chain B factor [Å2] | 22.9 |
| Avg water molecule B factor [Å2] | 35.7 |
| Avg ligand B factor (subunits A and B) [Å2] | 42.2, 54.2 |
| rms deviations from ideal geometry | |
| Covalent bond lengths [Å] | 0.008 |
| Bond angles [°] | 1.2 |
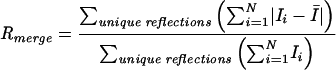 .
.
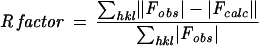 .
.
The free R factor is a cross-validation residual calculated by using 5% of the native data, which were randomly chosen and excluded from the refinement.
