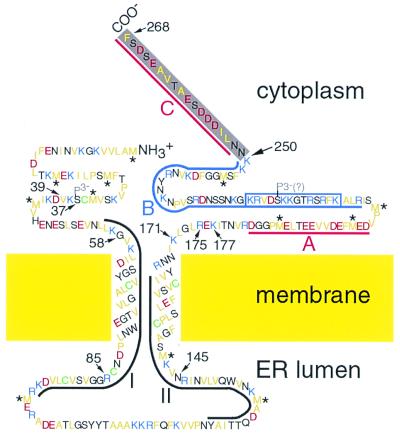
Figure 6.
Topological model of the TMV MP. The amino acid sequence was deduced from the ORF encoding the MP (32). Hydrophobic amino acid residues are yellow, basic residues are blue, acidic residues are red, and Cys residues are green. The trypsin-resistant core domain contains the first 249 or 250 residues, including the peptide (boxed) that was used to produce MP antibodies used in Figs. 2B and 5B. The C terminus (gray) was rapidly removed by trypsin. As previously defined (36), domains I (residues 56–96) and II (residues 125–164) are regions that are conserved among tobamovirus MPs and are outlined in black; domains A (residues 183–200) and C (residues 252–268) are acidic and are outlined in red; and domain B (residues 206–250) is basic and is outlined in blue. Cytoplasmic, transmembrane, ER luminal, and core domains were inferred from Western blots of proteinase K-treated microsomes, hydropathy analysis, fluorescence microscopy (16, 20, 22, 24), trypsin susceptibility, and MS. Transmembrane domains are presumed to be α-helical. Hydrophobic peptides (39–85 vs. 58–85) differed in length between the monomer and the core (Fig. 5), suggesting that the C terminus of the monomer may interact with its N terminus. Serine-37 is phosphorylated (P3-; ref. 37), and S 218 may be phosphorylated (P3-?) based on Prosite prediction and sequence conservation among tobamovirus MPs (not shown). Met residues (*) have been reported to generally be involved in protein–protein interactions (38).