Abstract
In Saccharomyces cerevisiae phosphatidylcholine (PC) is synthesized in the ER and transported to mitochondria via an unknown mechanism. The transport of PC synthesized by the triple methylation of phosphatidylethanolamine was investigated by pulsing yeast spheroplasts with l-[methyl-3H]methionine, followed by a chase with unlabeled methionine and subcellular fractionation. During the pulse, increasing amounts of PC and its mono- and dimethylated precursors (PMME and PDME, respectively) appear in similar proportions in both microsomes and mitochondria, with the extent of incorporation in microsomes being twice that in mitochondria. During the chase, the [3H]-methyl label from the precursors accumulates into PC with similar kinetics in both organelles. The results demonstrate that transport of methylated phospholipids from ER to mitochondria is 1) coupled to synthesis, 2) not selective for PC, 3) at least as fast as the fastest step in the methylation of PE, and 4) bidirectional for PMME and PDME. The interorganellar equilibration of methylated phospholipids was reconstituted in vitro and did not depend on ongoing methylation, cytosolic factors, ATP, and energization of the mitochondria, although energization could accelerate the reaction. The exchange of methylated phospholipids was reduced after pretreating both microsomes and mitochondria with trypsin, indicating the involvement of membrane proteins from both organelles.
INTRODUCTION
Phosphatidylcholine (PC) is a major lipid constituent of both mitochondrial membranes in yeast (Daum and Vance, 1997). Several mutant strains with defects in the biosynthesis of PC generate respiratory-deficient petites at high frequency (Griac et al., 1996), suggesting that PC is required for proper mitochondrial function. The enzymes catalyzing the biosynthesis of PC are localized outside the mitochondrion. Although the biosynthesis of PC and its regulation have been extensively characterized in yeast (Henry and Patton-Vogt, 1998), information on the mechanism of PC import by mitochondria, other than that it occurs (Daum et al., 1986; reviewed by Daum and Vance, 1997), remains scarce.
The triple methylation of phosphatidylethanolamine (PE) is the primary route for the synthesis of PC in yeast in the absence of exogenous choline (Carman and Zeimetz, 1996). The enzyme PE methyltransferase (PEMT) encoded by the CHO2 gene catalyzes the methylation of PE to phosphatidyl-N-monomethylethanolamine (PMME). Subsequently, the phospholipid methyltransferase (PLMT) encoded by the OPI3 gene methylates PMME to phosphatidyl-N,N-dimethylethanolamine (PDME) and PDME to PC (Kodaki and Yamashita, 1987; Summers et al., 1988; McGraw and Henry, 1989). S-adenosylmethionine (AdoMet) is the methyl donor in these reactions. Both methyltransferases were predicted to be integral membrane proteins (Kodaki and Yamashita, 1987) and have been localized to the ER membranes (Zinser et al., 1991). When choline is supplied in the growth medium, the CDP-choline pathway contributes to the net synthesis of PC (McMaster and Bell, 1994). It was recently shown that mitochondria do not preferentially incorporate PC from either of the biosynthetic pathways (Janssen et al., 1999).
In the present study the kinetics of the synthesis and transport to mitochondria of PC produced by methylation of PE have been characterized in S. cerevisiae, in pulse-chase experiments using l-[methyl-3H]methionine. Yeast spheroplasts were used instead of whole cells to enable immediate subcellular fractionation at the end of a labeling experiment by avoiding the time required for spheroplasting. Thus, the harsh conditions of mechanical disruption of the yeast cell wall yielding damaged organelles with high degrees of cross-contamination were avoided. It is shown that apart from mitochondrial import of PC, transport of the biosynthetic precursors of PC between ER and mitochondria occurs and that it is bidirectional. The interorganellar equilibration of methylated phospholipids was retained in vitro, in a reconstituted system of isolated mitochondria and microsomes. The implications of the findings for the formation of mitochondrial PC are discussed.
MATERIALS AND METHODS
Strains, Media, and Culture Conditions
Yeast strains used were D273-10B/A1 (MATα met6) lacking a functional homocysteine methyltransferase (Tzagoloff et al., 1976) and its parental wild-type D273–10B, SH414 (MATa his3 leu2 ura3 ade2 trp1 can1 opi3::URA3), SH458 (MATα his3 leu2 ura3 cho2::LEU2), SH922 (MATα his3 leu2 ura3 ade2 opi3::URA3 cho2::LEU2), and the congenic wild-type SH921 (MATα his3 leu2 ura3 ade2; Dowd et al., 2001; kind gifts of Dr. S.A. Henry, Cornell University, Ithaca, NY). The methionine auxotrophic strain D273-10B/A1 and strain D273-10B were grown at 30°C in semisynthetic medium containing 0.3% yeast extract (Sigma, St. Louis, MO), 0.1% glucose, 2% lactate as the carbon source, and various salts as described (Daum et al., 1982), under aerobic conditions. The strains SH414, SH458, SH922, and SH921 were grown in the same medium supplemented with 0.05 mM 2-(methylamino)-ethanol (Fluka, Buchs, Switzerland) and adenine, histidine, leucine, tryptophan, and uracil at a concentration of 20 mg/l each. The cells were harvested in the exponential growth phase at OD600 of 3–3.5 (Janssen et al., 2000). The methionine-free minimal medium used in the pulse-chase experiments contained, per liter: 6.7 g of yeast nitrogen base without amino acids (Difco, Detroit, MI), 20 g of lactate, 1 g of glucose, 20 mg of adenine, 20 mg of arginine, 20 mg of histidine, 60 mg of leucine, 30 mg of lysine, 300 mg of threonine, 20 mg of tryptophan, and 40 mg of uracil and was adjusted to pH 5.5.
Pulse-Chase Experiments
After washing the D273-10B/A1 cells with 1 mM EDTA, pH 7, spheroplasts were prepared as described (Daum et al., 1982) with the modification that the cells were treated with Zymolyase 100T (Seikagaku, Tokyo, Japan) at 0.5 mg/g cells (wet weight) in fresh growth medium adjusted to pH 7.4 and containing 0.7 M sorbitol (Reid, 1983). The spheroplasts were harvested, resuspended in 130 ml methionine-free minimal medium, 0.7 M sorbitol, 1% glucose, at a concentration of 0.1 g (wet weight) per ml, transferred to a 1-liter Erlenmeyer flask, and allowed to regenerate for 45 min in a water bath shaker at 30°C (Baker et al., 1988). During the last 15 min of the recovery period a flow of oxygen was bubbled through the suspension of spheroplasts.
At time zero, l-[methyl-3H]methionine (85 Ci/mmol, Amersham, Amersham, UK) and unlabeled l-methionine were added to the spheroplast suspension at final concentrations of 2.5 μCi/ml and 5 μM, respectively. After 5 min, the label was chased by adding unlabeled l-methionine from a freshly dissolved 0.23 M stock solution in minimal medium, to a final concentration of 25 mM. During the pulse-chase experiment, the spheroplast suspension was incubated at 30°C with gentle shaking and under oxygen bubbling as above.
At various time points samples were drawn from the spheroplast suspension. For the analysis of the spheroplast lipids, 0.5-ml samples were taken and immediately subjected to phospholipid extraction. For subcellular fractionation, 20-ml samples were drawn to which 0.1 ml 200 mM HgCl2 was added to inactivate the methyltransferases (Gaynor and Carman, 1990). After 2 min at room temperature the samples were chilled by dilution in 2 volumes of ice cold buffer S (1.2 M sorbitol, 20 mM KH2PO4, pH 7.4) also containing 1 mM HgCl2 and kept on ice until further processing, which was started within 40 min.
The spheroplasts were collected by centrifugation for 5 min at 1500 × g (SS34 rotor, Sorvall, Wilmington, DE). Excess Hg2+ was removed by resuspending the spheroplasts in buffer S also containing 0.1 mM EDTA and centrifugation as above. The spheroplasts were resuspended at a concentration of 1 g wet weight per 15 ml, in ice cold breakage buffer (0.6 M sorbitol, 10 mM MES, pH 6.0, 0.2% [wt/vol] BSA, 1 mM PMSF) containing 1 mM p-chloromercuriphenylsulfonic acid (PCMBS; Sigma) to inhibit the methyltransferases (Gaynor and Carman, 1990), and homogenized by 11 slow and forceful strokes in a glass Dounce homogenizer with a tight fitting pestle.
The procedure followed for subcellular fractionation is a shortened version of a published method (de Kroon et al., 1999). The homogenates were centrifuged for 5 min at 1500 × g to remove unbroken spheroplasts and nuclei. The supernatant was centrifuged for 12 min at 10,000 × g to pellet the mitochondria, which were further purified by discontinuous sucrose gradient centrifugation. The microsomal fraction was obtained as the 32,500 × g pellet of the 20,000 × g supernatant of the postmitochondrial supernatant. The subcellular fractions were frozen in liquid nitrogen and stored at -20°C until analysis. The purity of the subcellular fractions has been documented (de Kroon et al., 1999).
In Vitro Reconstituted System
Mitochondria loaded with [3H]-methyl-PMME were isolated from strain SH414. After harvesting, SH414 cells (∼40 g wet weight) were washed in minimal medium and incubated for 90 min at 30°C in 300 ml minimal medium with the above supplements, also containing 0.05 mM 2-(methylamino)-ethanol, and 1 mCi l-[methyl-3H]methionine. The cells were washed with 1 mM EDTA, spheroplasts were prepared, and mitochondria were isolated and purified by sucrose gradient centrifugation (de Kroon et al., 1999). Mitochondria for use in experiments involving energization were purified by Nycodenz gradient centrifugation (Janssen et al., 2002b). The final mitochondrial pellet was resuspended in 0.6 M sorbitol, 50 mM Tris, pH 7.5 (buffer C), also containing 0.5% (wt/vol) bovine serum albumin (BSA), frozen in aliquots in liquid nitrogen, and stored at -80°C until use.
Microsomes were isolated from strain D273-10B, unless indicated otherwise. The postmitochondrial supernatant (de Kroon et al., 1999) was centrifuged for 15 min at 21,500 × g to remove residual mitochondria. The supernatant was centrifuged for 30 min at 100,000 × g (Ti60 rotor, Beckman, Palo Alto, CA) to pellet the microsomes. To be able to separate the microsomes from the mitochondria after coincubation, the microsomes were pretreated as follows. The 100,000 × g pellet was resuspended at ∼10 mg protein per ml in buffer C, and a 7-ml aliquot was subjected to 9 cycles of 10 s ultrasonication at 0°C with 10-s intervals using a Branson sonifier 250 equipped with a microtip, operated at 60 W and 50% duty cycle. The suspension was loaded on 15% (vol/vol) Percoll (Pharmacia, Uppsala, Sweden) in buffer C and centrifuged for 15 min at 50,000 × g (SW28 rotor, Beckman). The upper 15 ml of the microsomal band was collected and centrifuged for 60 min at 170,000 × g (Ti60). The microsomal pellet was resuspended in buffer C and stored as aliquots at -80°C until use.
[3H]-methyl-PMME–labeled mitochondria and microsomes were coincubated in buffer C at concentrations of 1 and 2 mg protein/ml, respectively, in the presence of 5 mM AdoMet, at 30°C, unless indicated otherwise. At the time points indicated, samples (125 μl) were drawn and subjected to phospholipid extraction for further analysis. Alternatively, samples (500 μl) were applied to 10 ml 15% (vol/vol) Percoll in buffer C and centrifuged for 15 min at 50,000 × g (SW41 rotor, Beckman) to separate microsomes from mitochondria. Microsomes (1 ml) were collected from the top of the gradient and mitochondria (0.5 ml) from the bottom, with yields of 32 ± 8% and 48 ± 10% (±SD, n = 13) based on phospholipid phosphorus, respectively. After correcting for these yields, it was calculated that 100% of the lipid-associated [3H]-methyl label was recovered.
Before co-incubation, protease treatment of mitochondria and microsomes was carried out in buffer C at a protein concentration of 2 mg/ml with proteinase K or trypsin, at the concentrations indicated. After incubation for 30 min on ice, proteolysis was stopped by adding PMSF to a final concentration of 1 mM, or soy bean trypsin inhibitor (SBTI) in twofold excess (wt/wt) over trypsin, respectively.
Extraction and Analysis of Phospholipids
The lipids of spheroplasts and subcellular fractions were extracted as described (Bligh and Dyer, 1959). To quantitate the incorporation of the [3H]-methyl label into the lipids, the [3H]-methyl content of the lipid extracts was determined by liquid scintillation counting and related to the phospholipid phosphorus content of the lipid extracts determined by phosphate analysis (Rouser et al., 1970). To assess the distribution of the incorporated [3H]-methyl label over PC, PMME, and PDME, the lipid extracts were analyzed by one-dimensional high-performance TLC (HPTLC) on silica gel 60 (Merck, Darmstadt, FRG) using chloroform/methanol/25% ammonia, 70/25/4 (vol/vol/vol) as eluent. Subsequently, the radioactive spots on the TLC plate were quantitated by a Berthold Automatic TLC linear analyzer (Wildbad, FRG).
Other Methods
Yeast cytosol was isolated from strain D273-10B as described (Salama et al., 1993). The activity of Opi3p was assayed by incubating microsomes isolated from the cho2Δ strain SH458 with S-adenosyl-l-[methyl-3H]-methionine (Amersham) and determining the incorporation of the 3H label into chloroform soluble material as described (Janssen et al., 2002a). Protein concentrations were determined using the BCA method (Pierce, Rockford, IL) with 0.1% (wt/vol) SDS added and BSA as a standard. Immunoblot analysis was performed after SDS-PAGE on 9.75% gels as described (de Kroon et al., 1999).
RESULTS
Pulse-Chase Conditions for Yeast Spheroplasts
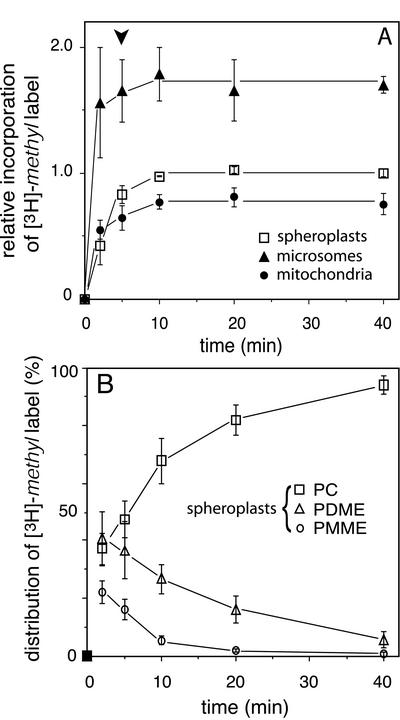
To monitor the time course of the appearance in mitochondria of newly synthesized PC, yeast spheroplasts from a met6 strain were pulsed for 5 min with [3H]methionine, followed by a chase with excess unlabeled methionine and subcellular fractionation. Under the conditions used, the extents of incorporation of the [3H]-methyl label into phospholipids were similar in spheroplasts and intact cells (our unpublished results), indicating that spheroplasts are a valid model for studying PC housekeeping in vivo. The incorporation of label in the lipids of spheroplasts stopped upon adding excess unlabeled methionine, resulting in a clear-cut chase (Figure 1A, squares). During a 5-min pulse, a typical level of incorporation of 2000 dpm [3H]-methyl label per nanomole of phospholipid phosphorus was obtained, which implies that some 18% of the [3H]-methyl label added to the medium was incorporated into the lipids. Ignoring the incorporation of the label into sterols (see below), this number was calculated to correspond to the single 3H methylation of 0.2 mol % of the spheroplast phospholipids.
Figure 1.
Pulse-chase experiments. (A) Time course of the relative incorporation of the [3H]-methyl label into the lipids of yeast spheroplasts, microsomes, and mitochondria. Because the amounts of label incorporated per nanomole of phospholipid phosphorus varied between experiments, the extents of incorporation in each individual experiment were normalized to the level of incorporation in spheroplasts during the chase (i.e., the mean value of the incorporation levels at t = 10, 20, and 40 min) before averaging (n = 3, ±SD). The arrow head indicates the start of the chase. (B) Distribution of the incorporated [3H]-methyl label over PMME, PDME, and PC in spheroplasts, presented as percentages of the total amount of label incorporated into the phospholipids (n = 3, ±SD).
Appearance of 3H-Methylated Phospholipids in Mitochondria
Figure 1 shows the results of pulse-chase experiments in which the incorporation of the [3H]-methyl label in the lipids of spheroplasts and derived subcellular fractions was examined. During the 5-min pulse rapid incorporation of the label was observed in the lipids of both the microsomal and the mitochondrial membranes to levels 1.7 and 0.8 times the level of incorporation in whole spheroplasts, respectively, consistent with the localization of the methyltransferases in the ER (Zinser et al., 1991). After introducing the chase, little further incorporation of label occurred, neither in the microsomes nor in the mitochondria, with the levels of incorporation in both organellar membranes remaining constant during the 35-min chase period (Figure 1A). This result shows that the appearance of 3H-methylated phospholipids in mitochondria was restricted to the time period of the pulse. The mitochondria-associated membrane (MAM), a subfraction of the ER that copellets with mitochondria, has been implicated in the transport of phosphatidylserine (PS) into mitochondria (Gaigg et al., 1995; Shiao et al., 1995). The time course and the level of incorporation of the [3H]-methyl label in the lipids of the MAM were similar as measured for the microsomes (our unpublished results).
Mono- and Dimethylated Intermediates
The incorporation of the [3H]-methyl label into the spheroplast lipids during a pulse-chase experiment was analyzed by HPTLC (Figure 1B). After the 5-min pulse considerable amounts of label were still present in PMME and PDME, which only gradually accumulated in the PC peak during the chase. At all time points, ∼5% of the lipid-incorporated [3H]-methyl label was recovered at the front. This probably reflected the incorporation of the label into sterols by the enzyme S-adenosylmethionine:Δ24-sterol-C-methyltransferase (Parks, 1978). The kinetics of the accumulation of the [3H]-methyl label in PC during pulse-chase experiments were similar in spheroplasts and intact cells, and in met6 and wild-type cells (our unpublished results) with the [3H]-methyl label-pulsed intermediates accumulating in PC with a half time of ∼10 min (Figure 1B).
The higher level of PDME compared with that of PMME that is observed throughout the time course of the pulse-chase experiments (Figure 1B) is explained by the Km of PLMT for PDME being more than twice that for PMME (Kodaki and Yamashita, 1989; Gaynor and Carman, 1990). Consistent with this observation, phospholipid extracts from wild-type yeast cells usually contain some 2% PDME, whereas PMME is hardly detectable (Summers et al., 1988; Daum et al., 1999).
Bidirectional Transport of PC Precursors between ER and Mitochondria
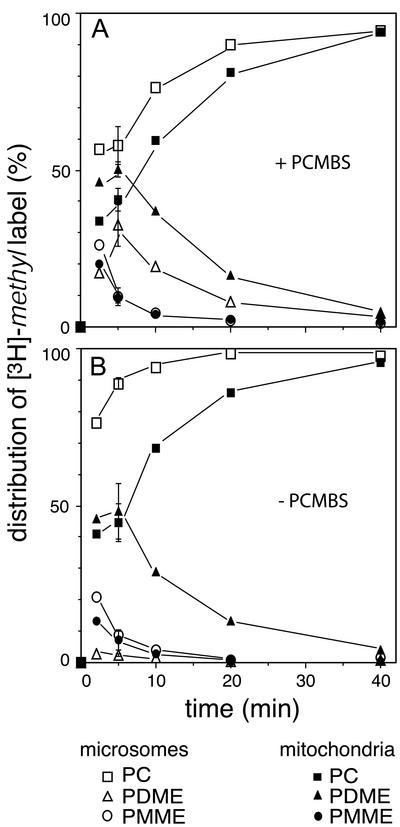
The distribution of the [3H]-methyl label over the methylated phospholipids in mitochondria and microsomes was analyzed during a pulse-chase experiment. The results (Figure 2A) unambiguously show that besides PC, its biosynthetic precursors PMME and PDME also appeared in the mitochondrial membranes. During the chase, the labeled intermediates gradually disappeared from the mitochondria and microsomes with similar kinetics. This result implies that mitochondrial PMME and PDME return to the ER for further methylation. In terms of the extent of methylation, the labeled lipids in the mitochondrial fraction were running slightly behind those in the microsomal fraction, as judged from the higher relative content of PDME (and the concomitant lower relative content of PC) in the mitochondria compared with that of the microsomes at all time points sampled (Figure 2A). This finding is most likely due to the inhibition of Opi3p by the methyltransferase inhibitor PCMBS (see below) not being immediate upon homogenization of the spheroplasts, leaving time for some further methylation of microsomal PDME.
Figure 2.
The rates of accumulation of the [3H]-methyl label in PC in mitochondria and microsomes, as observed after subcellular fractionation in the presence (A) and absence (B) of 1 mM PCMBS in the breakage buffer. The distribution of the [3H]-methyl label over PMME, PDME, and PC, is presented as a percentage of the total amount of label incorporated in the phospholipids of microsomes and mitochondria. The error bars at t = 5 min reflect the SD (n = 3). For experimental conditions see MATERIALS AND METHODS.
When the spheroplasts were fractionated in the absence of PCMBS, methylation resumed upon breaking the cells, as judged by the increased accumulation of [3H]-methyl label in PC in homogenized spheroplasts. Under these conditions, the time course of the accumulation of the label in PC in the mitochondria was hardly affected (cf. Figure 2, A and B, solid symbols). However, because of the resumption of methylation during subcellular fractionation, the accumulation of the label in PC in microsomes (and MAM, unpublished results) was found to be almost complete after 5 min of chase (Figure 2B). These results demonstrate that the intermediates in the mitochondria stay out of reach of the methyltransferases during the processing of the spheroplasts, providing further evidence that the intermediates PMME and PDME reside temporarily in the mitochondrial membranes and not in any adhering ER remaining. Furthermore, they exclude the possibility that the labeled lipids were redistributed during the processing of spheroplasts to subcellular fractions. The results of the pulse-chase experiments indicate that newly synthesized methylated phospholipids rapidly equilibrate between ER and mitochondria.
In Vitro Reconstitution of Transport of Methylated Phospholipids
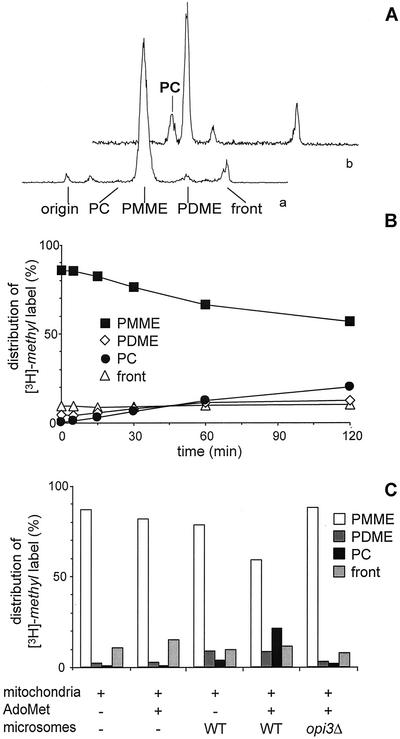
To investigate the mechanism of the equilibration of phospholipids between ER and mitochondria further, an in vitro system was set up. The temporary presence of PMME and PDME in mitochondria provides a novel opportunity for investigating interorganellar transport of PC and its precursors in vitro. On incubating mitochondria containing radiolabeled PC precursors with microsomes, the production of radiolabeled PC may serve as a measure for transport of the precursors from mitochondria to the microsomes. For this purpose, mitochondria were isolated from an opi3Δ strain that lacks the ability to convert PMME to PC, in order to preclude the complications arising from residual contamination with ER containing methyltransferase activity. An additional advantage of opi3Δ cells is that they are conveniently loaded with radiolabeled PMME by labeling with [3H]methionine, yielding mitochondria with >85% of the lipid-incorporated [3H]-methyl label present in PMME and no label in PC (Figure 3A, profile a; cf. Preitschopf et al., 1993; Janssen et al., 2002a).
Figure 3.
Mitochondrial [3H]-methyl-PMME is converted to [3H]-methyl-PC by microsomal PLMT in vitro in the presence of AdoMet. (A) Radioactivity scan of a TLC plate showing the distribution of the [3H]-methyl label in a lipid extract of opi3Δ mitochondria before (a) and after (b) incubation for 2 h with wild-type microsomes and AdoMet. (B) Time course of the conversion of [3H]-methyl-PMME into [3H]-methyl-PDME and [3H]-methyl-PC. The distribution of the incorporated [3H]-methyl label over PMME, PDME, and PC, and the front is presented as percentage of the total amount of label incorporated into lipids. Data are averaged from four independent experiments with the SD not exceeding 4%. (C) Distribution of the [3H]-methyl label over PMME, PDME, and PC, and the front after 2 h of incubation of [3H]-methyl-PMME-loaded opi3Δ mitochondria in the absence and presence of microsomes and AdoMet as indicated.
Incubation of the [3H]-methyl-PMME-loaded mitochondria in a mixture with microsomes from a wild-type strain (at a ratio of 1:2 based on protein content) and AdoMet, was sufficient for restoring the formation of PC as evidenced by the gradual conversion of 30% of [3H]-methyl-PMME into [3H]-methyl-PDME and [3H]-methyl-PC in 2 h (Figures 3A, profile b, and 3B). The formation of PC was strictly dependent on the addition of microsomes containing PLMT (Figure 3C). Without AdoMet added only some 10% of [3H]-methyl-PMME was methylated most likely because of endogenous AdoMet present (Figure 3C). During the 2 h of incubation no degradation of the labeled phospholipids occurred, because the recovery of the lipid-associated label was 100%, which is also reflected by the constant level of the relative amount of sterol-associated label running at the front (Figure 3B).
Exchange of [3H]-Methyl–labeled Lipids In Vitro
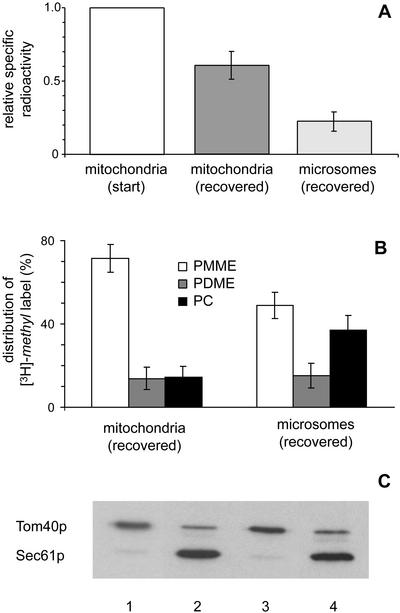
To determine whether the restoration of [3H]-methyl-PC synthesis depended on transport of [3H]-methyl-PMME to the microsomes, the localization of the produced [3H]-methyl labeled PC was assessed. After 2 h of incubation with microsomes and AdoMet, the mitochondria had lost about one third of their initial content of [3H]-methyl labeled lipids, which was recovered in the microsomal fraction (Figure 4A). HPTLC analysis of the lipid distribution of the [3H]-methyl label in the two fractions revealed that the [3H]-methyl-PC produced was not only present in the microsomes, but also in the mitochondria (Figure 4B). Compared with the mitochondrial fraction, the microsomal fraction contained relatively more label in PC at the expense of PMME. Immunoblot analysis of the purity of the recovered fractions compared with the starting material (Figure 4C) demonstrated that cross-contamination between the fractions was minor and did not account for the observed redistribution of [3H]-methyl labeled lipids between mitochondria and microsomes. The results from the in vitro reconstituted system reflect exchange of PC and its precursors between mitochondria and microsomes, similar to that observed in the in vivo pulse-chase experiments, albeit at a slower rate.
Figure 4.
Mitochondrial [3H]-methyl-PMME and its methylation products are exchanged between mitochondria and microsomes during coincubation. (A) Distribution of the [3H]-methyl label between mitochondria and microsomes that were separated by Percoll gradient centrifugation after incubation for 2 h of [3H]-methyl-PMME-loaded opi3Δ mitochondria with wild-type microsomes and AdoMet. The specific radioactivities, defined as dpm 3H per nanomole of phospholipid phosphorus, have been normalized to that of the mitochondria at time zero. (B) Distribution of the [3H]-methyl label over PMME, PDME, and PC, shown as the percentages of the total amounts of label incorporated into the phospholipids of mitochondria and microsomes after coincubation as above. (C) Samples corresponding to 1 μg mitochondria (lanes 1 and 3) and 3 μg microsomes (lanes 2 and 4; protein-based) before (lanes 1 and 2) and after (lanes 3 and 4) coincubation and separation as above were subjected to SDS-PAGE, blotted onto nitrocellulose, and immunostained with antibodies against Tom40p and Sec61p. The error bars in A and B represent the SD (n = 15).
Mechanism of Exchange
Replacing the wild-type microsomes by opi3Δ, cho2Δ, or cho2Δ opi3Δ microsomes in the reconstituted system did not affect the intermembrane transfer of radiolabeled lipids, demonstrating that the exchange in vitro does not require the activity or the presence of the methyltransferases (our unpublished results). Using the methylation of [3H]-methyl-PMME as a measure for its intermembrane transport, the effect of several other factors on the process was investigated. The addition of cytosol up to a concentration of 12.5 mg protein per ml did not have any effect. Table 1 summarizes the effects of energizing the mitochondria and of introducing ATP in the reconstituted system. The addition of the respiratory substrate ethanol significantly enhanced the efficiency of methylation. This effect disappeared in the presence of the uncoupler p-trifluoromethoxycarbonyl cyanide phenylhydrazine (FCCP), suggesting that the proton motive force across the mitochondrial inner membrane promotes the transfer of PMME to the microsomes. For reasons presently not understood, the enhanced methylation was not observed when NADH was used as respiratory substrate (Table 1). The addition of ATP (or GTP, unpublished results), irrespective of the presence of cytosol (our unpublished results), slightly enhanced methylation in the absence of Mg2+, whereas with Mg2+ present methylation was reduced (Table 1). Because Mg2+ was reported to stimulate the activity of Opi3p (Gaynor and Carman, 1990), the inhibition of methylation observed here must be due to Mg2+ affecting the transfer of [3H]-methyl-PMME.
Table 1.
Effect of respiratory substrates and ATP/Mg2+ on the methylation of mitochondrial [3H]-methyl-PMME in the in vitro reconstituted system
| Additive(s) | Relative methylation of [3H]-methyl-PMME |
|---|---|
| None | 1.0 |
| Ethanol (17 mM) | 1.24 ± 0.05 |
| Ethanol (17 mM) + FCCP (10 μM) | 1.04 ± 0.05 |
| NADH (2 mM) | 1.0 ± 0.05 |
| NADH (2 mM) + FCCP (10 μM) | 1.0 ± 0.05 |
| ATP (2 mM) | 1.15 ± 0.07 |
| ATP (2 mM) + MgCl2 (5 mM) | 0.7 ± 0.2 |
| MgCl2 (5 mM) | 0.6 ± 0.2 |
Mitochondria from the opi3Δ strain and wild-type microsomes were incubated for 2 h at 1 mg/ml each in the presence of AdoMet and the additives indicated, as detailed in MATERIALS AND METHODS. When ethanol or NADH was added, the buffer also contained 5 mM Pi to maintain mitochondrial intactness (Janssen et al., 2002b). FCCP was added from a 10 mM stock solution in ethanol. Methylation was quantified as the increase in the percentage of [3H]-methyl label present in PDME and PC upon 2 h of incubation and was normalized to that in the absence of additives (32 ± 5% of the lipid-incorporated [3H]-methyl label). Results are mean ± SD (n = 3).
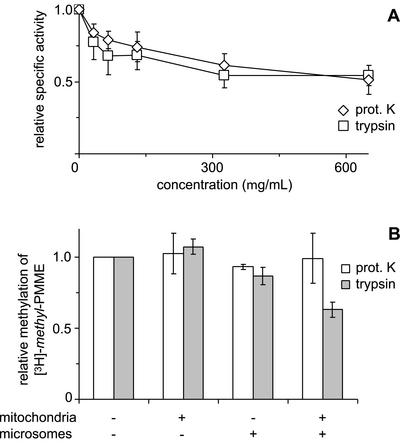
To investigate whether surface-exposed mitochondrial and/or ER proteins are involved in the intermembrane transfer of PMME, mitochondria and microsomes were treated with proteases before coincubation with AdoMet. First, the effect of protease treatment on Opi3p enzyme activity was assessed using microsomes from a cho2Δ strain. After incubation with increasing concentrations of proteinase K or trypsin up to 650 μg/ml, at least 50% of the methylation activity of Opi3p was retained (Figure 5A). Under these conditions, the enzyme activity of Cho2p measured in opi3Δ microsomes was completely abolished (our unpublished results). Pretreatment of mitochondria, microsomes, or both with proteinase K (500 μg/ml) did not interfere with the methylation of mitochondrial [3H]-methyl-PMME in the reconstituted system (Figure 5B). Pretreatment with trypsin up to 500 μg/ml of mitochondria or microsomes did not significantly reduce methylation either (shown for 100 μg/ml trypsin in Figure 5B). However, after preincubating both organelles with trypsin, methylation was reduced by 40 and 60% at 100 μg/ml (Figure 5B) and 500 μg/ml trypsin, respectively. These data indicate that surface-exposed proteins that are present on both organelles can mediate the exchange. Furthermore, they show that the transfer of mitochondrial [3H]-methyl-PMME to the microsomes rather than the Opi3p-catalyzed reaction is rate limiting for methylation.
Figure 5.
Effects of treatment with proteases on (A) the enzyme activity of Opi3p and (B) the methylation of mitochondrial [3H]-methyl-PMME in the reconstituted system. (A) Microsomes from a cho2Δ strain were incubated with the indicated concentrations of proteinase K or trypsin before incubation with [methyl-3H]AdoMet to assay enzyme activity. TLC analysis revealed that >90% of the lipid-incorporated label ended up in PC over the entire range of protease concentrations tested. Data have been normalized to the specific activity of Opi3p in mock-treated microsomes (n = 3, ±SD). (B) Mitochondria from the opi3Δ strain and/or wild-type microsomes were treated with proteinase K (500 μg/ml) or trypsin (100 μg/ml), as indicated, before coincubation at 1 mg/ml each in the presence of AdoMet. Methylation was quantified as the increase in the percentage of [3H]-methyl label present in PDME and PC upon 2 h of incubation and normalized to that of the control. Data are mean values from two independent experiments, with the error bars representing the variation.
DISCUSSION
The appearance in mitochondria of PC, newly synthesized by the methylation route, has been studied in vivo, in pulse-chase experiments on yeast spheroplasts, and in vitro, in a reconstituted system consisting of isolated mitochondria and microsomes. The results of both experimental approaches demonstrate rapid equilibration of PC as well as of its biosynthetic precursors between ER membranes and mitochondria. In the forthcoming, the results and their implications will be discussed in the light of the generally accepted transport model, in which PC after its synthesis in the ER is imported by mitochondria.
Subcellular fractionation of pulsed-chased spheroplasts revealed that the appearance of newly synthesized [3H]-methyl labeled phospholipids in the mitochondrial membranes is strictly coupled to ongoing methylation. After introducing the chase with cold methionine, no further increase of the mitochondrial content of 3H-labeled lipids at the expense of microsomal 3H-labeled lipids was observed (Figure 1). The apparent coupling between the methylation of PE and the appearance of the reaction products in the mitochondria is underscored by the similar kinetics of label accumulation in PC observed in mitochondria and microsomes (Figure 2A). The results obtained in the absence of PCMBS (Figure 2B), where the methyltransferases resume activity during the processing of the spheroplasts, unequivocally prove that the intermediates are present in the mitochondrial membranes at the times of sampling rather than in any adhering ER, as they stay out of reach of (reactivated) PLMT, which is localized in the ER (Zinser et al., 1991).
The implications of the present results for the formation of mitochondrial PC are several. It is generally accepted that mitochondria acquire PC by import from the ER (Daum and Vance, 1997; Voelker, 2000). In this light the data show that net intermembrane transport is coupled to synthesis and proceeds at least as fast as the fastest step in the methylation of PE. Transport is not selective as both PC and the intermediates are imported by mitochondria to extents reflecting their relative contents in the ER. The bidirectionality of the transport process is evidenced by the gradual disappearance of PMME and PDME from the mitochondria during the chase. Degradation of substantial amounts of the intermediates in the mitochondria is ruled out as the [3H]-methyl content of the mitochondrial lipids stays at a constant level during the chase. Combined, these results infer the immediate return to the mitochondrion of the labeled intermediates after further methylation in ER or MAM, supporting the existence of continuous exchange of methylated phospholipids between ER and mitochondria.
The most probable mechanism for the bidirectional continuous transport is via membrane contacts between ER and mitochondrial outer membrane, which may involve the MAM (Gaigg et al., 1995). A pulse-labeling study in CHO cells has provided evidence that PS traverses the MAM on its way from ER to mitochondria (Shiao et al., 1995). However, based on the present results, no special role in mitochondrial PC import can be assigned to MAM.
An alternative explanation for the cosynthetic appearance of PMME, PDME, and PC in the mitochondrial membranes does not involve intermembrane transport. Instead, PEMT and PLMT would be able to methylate phospholipid substrates present in a neighboring (“trans”) membrane. The numerous contacts between ER/MAM and mitochondrial outer membrane (Daum and Vance, 1997; Rizzuto et al., 1998) would provide the sites where the ER membrane proteins PEMT and PLMT have access to substrate molecules residing in the mitochondrial outer membrane. This trans-catalysis hypothesis was proposed based on the cooperative activity of PEMT and PLMT observed in mixtures of microsomes from cho2 and opi3 disruptant strains (Janssen et al., 2002a). Interestingly, the PE methyltransferase of Rhodobacter sphaeroides is a soluble enzyme (Cain et al., 1984; Arondel et al., 1993), which is active when associated with the membrane. The yeast methyltransferases might have retained this property and be catalytically active on a “trans” membrane.
The redistribution of PMME between mitochondria and microsomes established in the in vitro system, argues against transcatalysis as exclusive explanation for the parallel appearance and disappearance of PMME and PDME in mitochondria and microsomes observed in spheroplasts. The transfer of PMME to the microsomes and the subsequent return to the mitochondria of part of the PC (and PDME) produced are in agreement with the bidirectional continuous exchange of methylated phospholipids inferred by the pulse-chase experiments. The rate of intermembrane exchange of phospholipids in the reconstituted system was significantly slower than that observed in the pulse-chase experiments. This is probably due to the lack of synthesis of new lipid molecules in vitro, and thus of membrane growth.
The transfer in vitro of PMME to the microsomes was independent of the presence of the methyltransferases. It did not require cytosolic factors, nor was it greatly affected by the presence of ATP, as was found previously for mitochondrial import of PS and export of PE in vitro (Achleitner et al., 1999). The process was stimulated when the mitochondria were energized by supplying ethanol as respiratory substrate. Although treatment of the donor or the acceptor membrane with trypsin had no effect, treatment of both donor and acceptor reduced the transfer of PMME to the microsomes by a factor of 2 (Figure 5B). These results strongly suggest that surface-exposed proteins present on both organellar membranes can mediate the exchange of PMME via intermembrane contacts. These proteins appear to be highly resistant to inactivation by trypsin, because it is impossible to completely abolish the transfer. Proteolysis with proteinase K did not affect PMME transfer, whereas this pretreatment was reported to partially inhibit the in vitro transport of PS from MAM to mitochondria (Achleitner et al., 1999), suggesting that import of PS and export of PMME use different routes.
In terms of efficiency, the in vitro reconstituted system reported here compares favorably to other reconstituted phospholipid transport systems described in the literature. Moreover, this is the first in vitro demonstration that the PC formed is localized to both microsomal and mitochondrial membranes. Previously, the export of PE from mitochondria to ER was reconstituted in permeabilized yeast cells and in crude mitochondria with MAM still attached (Achleitner et al., 1995, 1999), using the conversion of PE into PC as criterion for transport. In both systems, this process proceeded relatively inefficiently with <10% of labeled PE from the mitochondria being converted into PDME and PC in 90 min (Achleitner et al., 1999).
In the continuous exchange of (methylated) phospholipids at sites of contact between the mitochondrial outer membrane and the ER, the phospholipids distribute according to their concentration gradients. In the process, the characteristic phospholipid compositions of the ER and the mitochondrial membranes might be maintained by the specific localizations of the phospholipid biosynthetic enzymes involved. Contrary to PS synthase, the methyltransferases are reportedly not enriched in the MAM (Gaigg et al., 1995), whereas PS decarboxylase 1 is localized at the mitochondrial inner membrane (Zinser et al., 1991). This distribution ensures that PE is enriched in mitochondria and that the PC content of the ER is higher than that of mitochondria (Janssen et al., 2000). The challenge now is to identify the proteins mediating intermembrane phospholipid flow between mitochondria and ER.
Acknowledgments
We thank Dr. M.J.F.W. Janssen for helpful discussions and Drs. R. Lill and R. Schekman for providing antibodies.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-08-0460. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-02-0460.
Abbreviations used: MAM, mitochondria associated membranes; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PDME, phosphatidyldimethylethanolamine; PMME, phosphatidylmonomethylethanolamine; PS, phosphatidylserine; PCMBS, p-chloromercuriphenylsulfonic acid; HPTLC, high performance TLC.
References
- Achleitner, G., Zweytick, D., Trotter, P.J., Voelker, D.R., and Daum, G. (1995). Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 270, 29836–29842. [DOI] [PubMed] [Google Scholar]
- Achleitner, G., Gaigg, B., Krasser, A., Kainersdorfer, E., Kohlwein, S.D., Perktold, A., Zellnig, G., and Daum, G. (1999). Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264, 545–553. [DOI] [PubMed] [Google Scholar]
- Arondel, V., Benning, C., and Somerville, C.R. (1993). Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J. Biol. Chem. 268, 16002–16008. [PubMed] [Google Scholar]
- Baker, D., Hicke, L., Rexach, M., Schleyer, M., and Schekman, R. (1988). Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell 54, 335–344. [DOI] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Cain, B.D., Donohue, T.J., Shepherd, W.D., and Kaplan, S. (1984). Localization of phospholipid biosynthetic enzyme activities in cell-free fractions derived from Rhodopseudomonas sphaeroides. J. Biol. Chem. 259, 942–948. [PubMed] [Google Scholar]
- Carman, G.M., and Zeimetz, G.M. (1996). Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271, 13293–13296. [DOI] [PubMed] [Google Scholar]
- Daum, G., Böhni, P.C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033. [PubMed] [Google Scholar]
- Daum, G., Heidorn, E., and Paltauf, F. (1986). Intracellular transfer of phospholipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 878, 93–101. [DOI] [PubMed] [Google Scholar]
- Daum, G., and Vance, J.E. (1997). Import of lipids into mitochondria. Prog. Lipid Res. 36, 103–130. [DOI] [PubMed] [Google Scholar]
- Daum, G. et al. (1999). Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast 15, 601–614. [DOI] [PubMed] [Google Scholar]
- de Kroon, A.I.P.M., Koorengevel, M.C., Goerdayal, S.S., Mulders, P.C., Janssen, M.J.F.W., and De Kruijff, B. (1999). Isolation and characterization of highly purified mitochondrial outer membranes of the yeast Saccharomyces cerevisiae (method). Mol. Membr. Biol. 16, 205–211. [DOI] [PubMed] [Google Scholar]
- Dowd, S.R., Bier, M.E., and Patton-Vogt, J.L. (2001). Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J. Biol. Chem. 276, 3756–3763. [DOI] [PubMed] [Google Scholar]
- Gaigg, B., Simbeni, R., Hrastnik, C., Paltauf, F., and Daum, G. (1995). Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta 1234, 214–220. [DOI] [PubMed] [Google Scholar]
- Gaynor, P.M., and Carman, G.M. (1990). Phosphatidylethanolamine methyltransferase and phospholipid methyltransferase activities from Saccharomyces cerevisiae. Enzymological and kinetic properties. Biochim. Biophys. Acta 1045, 156–163. [DOI] [PubMed] [Google Scholar]
- Griac, P., Swede, M.J., and Henry, S.A. (1996). The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J. Biol. Chem. 271, 25692–25698. [DOI] [PubMed] [Google Scholar]
- Henry, S.A., and Patton-Vogt, J.L. (1998). Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Progr. Nucleic Acid Res. Mol. Biol. 61, 133–177. [DOI] [PubMed] [Google Scholar]
- Janssen, M.J.F.W., Koorengevel, M.C., De Kruijff, B., and De Kroon, A.I.P.M. (1999). Transbilayer movement of phosphatidylcholine in the mitochondrial outer membrane of Saccharomyces cerevisiae is rapid and bi-directional. Biochim. Biophys. Acta 1421, 64–76. [DOI] [PubMed] [Google Scholar]
- Janssen, M.J.F.W., Koorengevel, M.C., De Kruijff, B., and De Kroon, A.I.P.M. (2000). The PC/PE ratio in the membranes of Saccharomyces cerevisiae varies with the growth phase. Yeast 16, 641–650. [DOI] [PubMed] [Google Scholar]
- Janssen, M.J.F.W., De Jong, H.M., De Kruijff, B., and De Kroon, A.I.P.M. (2002a). Cooperative activity of phospholipid-N-methyltransferases localized in different membranes. FEBS Lett. 513, 197–202. [DOI] [PubMed] [Google Scholar]
- Janssen, M.J.F.W., De Kruijff, B., and De Kroon, A.I.P.M. (2002b). Phosphate is required to maintain the outer membrane integrity and membrane potential of respiring yeast mitochondria. Anal. Biochem. 300, 27–33. [DOI] [PubMed] [Google Scholar]
- Kodaki, T., and Yamashita, S. (1987). Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262, 15428–15435. [PubMed] [Google Scholar]
- Kodaki, T., and Yamashita, S. (1989). Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur. J. Biochem. 185, 243–251. [DOI] [PubMed] [Google Scholar]
- McGraw, P., and Henry, S.A. (1989). Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster, C.R., and Bell, R.M. (1994). Phosphatidylcholine biosynthesis via the CDP-choline pathway in Saccharomyces cerevisiae. Multiple mechanisms of regulation. J. Biol. Chem. 269, 14776–14783. [PubMed] [Google Scholar]
- Parks, L.W. (1978). Metabolism of sterols in yeast. Crit. Rev. Microbiol. 6, 301–341. [DOI] [PubMed] [Google Scholar]
- Preitschopf, W., Lückl, H., Summers, E., Henry, S.A., Paltauf, F., and Kohlwein, S.D. (1993). Molecular cloning of the yeast OPI3 gene as a high copy number suppressor of the cho2 mutation. Curr. Genet. 23, 95–101. [DOI] [PubMed] [Google Scholar]
- Reid, G.A. (1983). Pulse labeling of yeast cells and spheroplasts. Methods Enzymol. 97, 324–329. [DOI] [PubMed] [Google Scholar]
- Rizzuto, R., Pinto, P., Carrington, W., Fay, F.S., Fogarty, K.E., Lifshitz, L.M., Tuft, R.A., and Pozzan, T. (1998). Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766. [DOI] [PubMed] [Google Scholar]
- Rouser, G., Fleischer, S., and Yamamoto, A. (1970). Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496. [DOI] [PubMed] [Google Scholar]
- Salama, N.R., Yeung, T., and Schekman, R.W. (1993). The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 12, 4073–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao, Y.J., Lupo, G., and Vance, J.E. (1995). Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270, 11190–11198. [DOI] [PubMed] [Google Scholar]
- Summers, E.F., Letts, V.A., McGraw, P., and Henry, S.A. (1988). Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics 120, 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff, A., Akai, A., and Foury, F. (1976). Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 65, 391–395. [DOI] [PubMed] [Google Scholar]
- Voelker, D.R. (2000). Interorganelle transport of aminophospholipids. Biochim. Biophys. Acta 1486, 97–107. [DOI] [PubMed] [Google Scholar]
- Zinser, E., Sperka-Gottlieb, C.D.M., Fasch, E.V., Kohlwein, S.D., Paltauf, F., and Daum, G. (1991). Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]