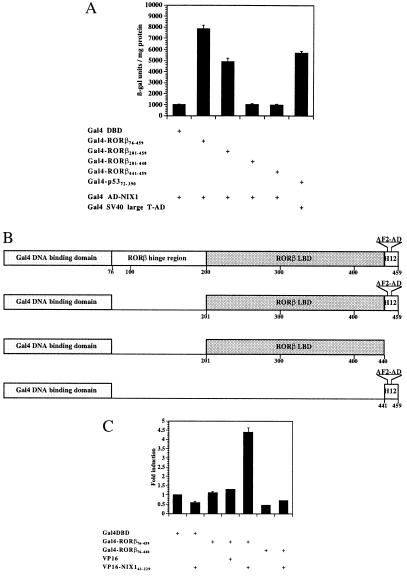
Figure 2.
Functional interaction between NIX1 and RORβ in vivo. (A) NIX1 interacts with RORβ in an AF2-D-dependent fashion. Plasmids expressing different deletion mutants of RORβ fused to the Gal4 DBD were introduced into the yeast reporter strain SFY526 together with NIX1 fused to the Gal4 activation domain. The transformants were grown in liquid dropout medium lacking tryptophan and leucine. Extracts were prepared and assayed for β-galactosidase activity, which is expressed in units per mg of protein. The stimulation in β-galactosidase activity resulting from the in vivo interaction between the Gal4-RORβ constructs and the NIX1-AD is indicated. Interaction between Gal4-p5373–708 and simian virus 40 large-T-AD served as an internal positive control. (B) Schematic representation of different RORβ deletion mutants fused to the Gal4 DBD domain. The AF2-D core (AF2-AD) motif is located at amino acid residues 445–451 within helix 12 (H12). (C) RORβ requires the AF2-D core for the interaction with NIX1 in mammalian cells. (GALp)3-TK LUC (1.25 μg) was cotransfected into CV1 cells with the indicated expression plasmids (200 ng each) coding for Gal4 DBD, Gal4 DBD fused to various RORβ deletion mutants, VP16 activation domain, and VP16-NIX1 amino acids 61–229. Fold induction averaged from seven independent experiments indicates the relative luciferase activity of the Gal4 DBD fusion proteins alone or in combination with the VP16 constructs over the Gal4 DBD vector.