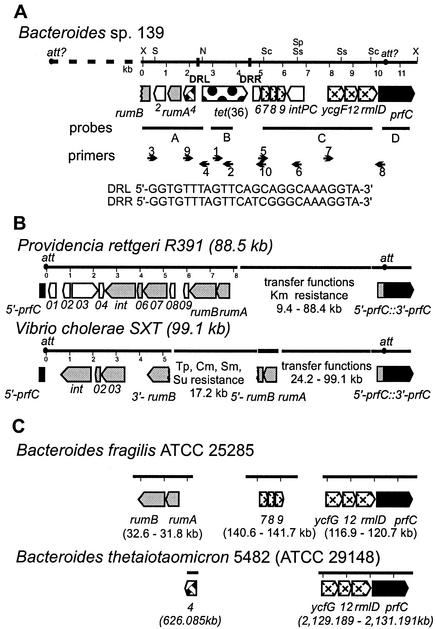
FIG. 2.
(A) Schematic diagram showing the organization of the tet(36) region from Bacteroides sp. strain 139. The numbers show the distance in kilobases from the leftmost XbaI site to the rightmost SphI site. Restriction sites shown are those of XbaI (X), SalI (S), NsiI (N), ScaI (Sc), SphI (Sp), and SstI (Ss). The major potential genes and their respective orientations are represented by arrows. Direct repeats flanking tet(36) are represented by filled boxes, and the sequences of the direct repeat upstream of tet(36) (DRL) and downstream of tet(36) (DRR) are shown. Probes used in Southern and dot blot analyses are represented by horizontal lines below the tet(36) region. Small arrows indicate the positions and directions of primers used for PCR analyses. (B) Schematic diagrams of related CTn-like elements R391 from P. rettgeri and SXT from V. cholerae are shown below the tet(36) region. (C) A schematic diagram of the organization and positions of genes from the B. fragilis type strain ATCC 25285 genome and the B. thetaiotaomicron type strain 5482 (ATCC 29148), which encode proteins homologous to those encoded by genes from the tet(36) region, is shown. ORFs are defined as follows: homologous amino acid sequences related to those present in both the SXT element and R391 are indicated by gray arrows, nonhomologous element sequences are indicated by unfilled arrows, the tet(36) gene sequence is indicated by a spotted arrow, homologous sequences not present in either the SXT element or R391 are indicated by hatched arrows, and prfC gene sequences are indicated by black arrows. orf4 (weave) is also found in B75482. Dashed lines indicate regions of the elements not drawn to scale. Antibiotic resistances are indicated as follows: kanamycin (Km), trimethoprim (Tp), chloramphenicol (Cm), streptomycin (Sm), and sulfonamides (Su).