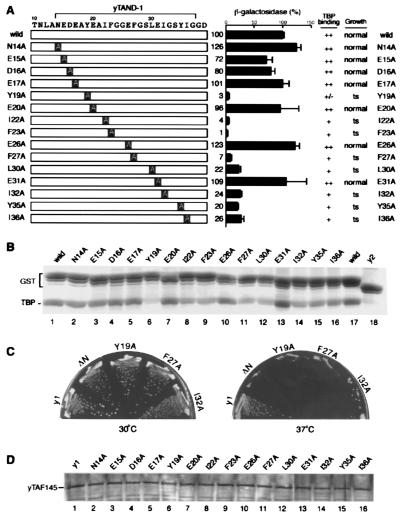
Figure 4.
The interaction of yTAND-1 with the concave surface of TBP is essential for its functions. (A) Alanine-scanning substitution mutants of yTAND-1 used in this study are schematically presented. These mutants were used for TBP-binding, growth, and transcription assays, and the results are summarized. For transcription analysis, indicated fragments were expressed in yeast as fusions with the GAL4 DNA-binding domain and tested for GAL4-dependent transcription, as described in the Fig. 3 legend. Note that although the point mutations within the yTAF145 gene caused only subtle effects, more obvious, but correlated, effects were observed when the point mutations were introduced into the yTAF145 gene in which TAND-2 (43–71 aa) had been internally deleted. The results with the gene background lacking TAND-2 are represented in this figure. (B) TBP-binding activity of the yTAND-1 mutants. The yTAND-1 mutants were fused to yTAND-2 and expressed in E. coli as GST-tagged proteins. Interaction of these fusions with TBP was determined as described in the Fig. 1 legend, and a Coomassie blue-stained gel is represented. (C) Growth phenotype of yeast carrying a defined mutation in yTAF145 gene. The yTAF145 genes harboring alanine-scanning mutations were introduced into yeast, replacing the wild-type gene by plasmid shuffling. After plasmid shuffling, the resulting strains were grown on yeast extract/peptone/dextrose plates at 30°C (Left) and 37°C (Right) for 3 days. Results are summarized in A, and photographs of yeast plates represent the selected mutants. Strains ΔN and y1 express yTAF145 proteins lacking the whole TAND (6–96 aa) and TAND-2 (43–71 aa), respectively (D). Expression level of the yTAF145 mutants in yeast. Yeast strains harboring yTAF145 mutants were grown at 25°C and then shifted to 37°C. Cells were harvested 24 h after the temperature shift and tested for expression level of yTAF145 mutants by immunoblotting.