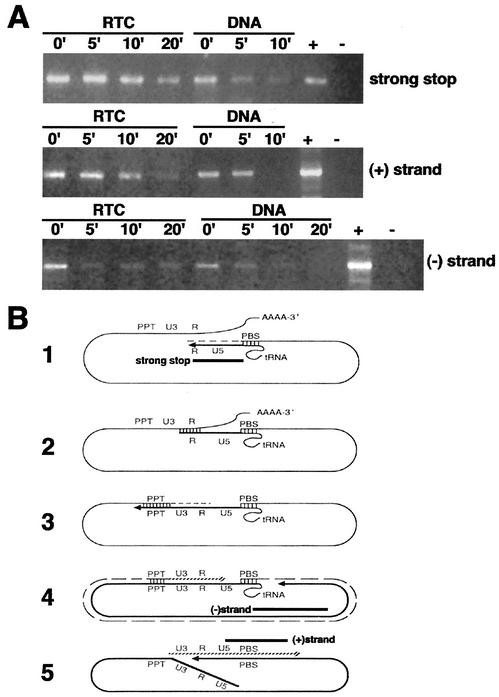
FIG. 7.
The viral DNA within the RTC is partially protected from micrococcal nuclease digestion. (A) Equilibrium density fractions containing the peak of the viral DNA (4 h postinfection) were incubated in the presence of micrococcal nuclease and 1.6 mM CaCl2. The reactions were stopped by addition of 4 mM EGTA at the indicated time points and analyzed by PCR with primers specific for negative-strand, positive-strand, and strong-stop DNAs. Naked viral DNA was prepared from the same density fractions by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation and then incubated as described above in the presence of micrococcal nuclease. −, No DNA; +, HIV-1 plasmid DNA. (B) Schematic representations of reverse transcription. The regions of the genome amplified by PCR in panel A are indicated by black bars. Diagram 1 shows the synthesis of the negative-strand strong-stop DNA starts at the primer-binding site (PBS), where a partially unfolded tRNA is bound. RNase H degrades the positive-strand RNA template so that the first strand transfer can take place. In diagram 2, a bridge is formed between the two complementary R sequences, and RT can jump on either of the RNA strands to elongate the negative-strand DNA in diagram 3. The RNA template is degraded except for the PPT. In diagram 4, the synthesis of the negative strand is completed. In diagram 5, the synthesis of the positive-strand strong-stop DNA starts at the PPT. The tRNA primer is removed, and the two strong-stop strands pair, forming a circular molecule suitable for elongation of the positive strand. At the end of the elongation process, the circular intermediate is opened into a linear double-stranded DNA molecule.