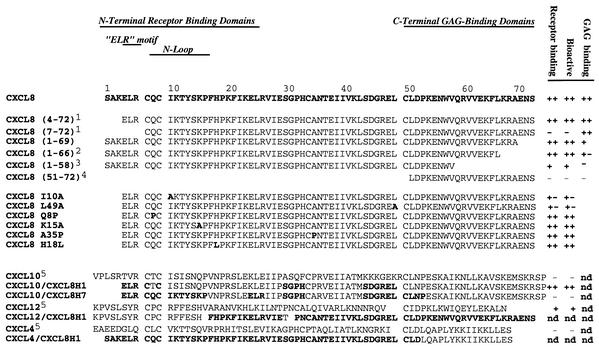
FIG. 1.
Amino acid sequences of the following hybrid analogs used in this study: CXCL8 (8, 9, 10, 15), CXCL8 residues 4 to 72 (8, 9, 10, 15, 19), CXCL8 residues 7 to 72, (8, 15, 24), CXCL8 residues 1 to 69 (8, 24), CXCL8 residues 1 to 66 (14, 18, 24), CXCL8 residues 1 to 58 (8, 24), CXCL8 residues 51 to 72 (8, 24), CXCL8 I10A (10, 19), CXCL8 L49A (10), CXCL8 Q8P (9), CXCL8 K15A (9), CXCL8 A35P (9), CXCL8 H18L (9), CXCL10 (9), CXCL10-CXCL8H1 (9), CXCL10-CXCL8H7 (9), CXCL12 (4), CXCL12-CXCL8H1, CXCL4 (4), and CXCL4-CXCL8H1. The first group comprises the analogs with either N- or C-terminal truncation. The analogs in the second group are analogs of CXCL8 with substitutions (substituted residues are shown in boldface type). The third group contains hybrid analogs with CXCL8 residues shown in boldface type. 1, analysis of 1H nuclear magnetic resonance parameters has shown that the core structure is the same as that for WT CSCL8, thus differences in binding and function cannot be attributed to incorrect folding of the protein (19); 2, unable to bind vCKBP-1 (14); 3, able to bind neutrophils and induce chemotaxis but with 30 times less potency (8); 4, able to restore bioactivity to CXCL8 residues 1 to 51 (18); 5, CXCL12 and CXCL4 are unable to bind vCKBP-3 and CXCL10 can bind vCKBP-3 but with reduced affinity (Table 1); ND, not determined; +, medium; ++, high; −, undetectable; +−, low.