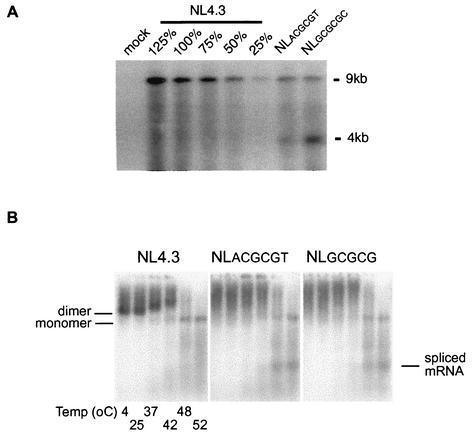
FIG. 3.
(A) Mutations in DIS stem-loop inhibit virion packaging of genomic RNA. Virion particles were produced by transfecting the indicated proviral DNA into 293T cells. Virion RNA samples that were normalized via a quantitative p24 assay were separated by electrophoresis on a denatured agarose gel. A dilution series of wt virion RNA was used to construct a standard curve. The impact of mutations on virion RNA packaging was visualized by Northern analysis and quantified by phosphorimaging. Results are representative of three sets of experiments. (B) The conformation but not the stability of HIV-1 virion RNA dimers from transfected 293T cells was affected by mutations of the DIS stem-loop. Virion RNA dimers were isolated from transfected 293T cells and prepared as described in Materials and Methods. Viral monomeric and dimeric RNA species were separated on a 1% native agarose gel via electrophoresis after the samples were heated for 10 min at various temperatures (4, 25, 37, 42, 48, and 52°C, lanes 1 to 6, respectively). Viral RNAs were visualized by Northern analysis using a radioactive riboprobe that specifically recognizes HIV-1 RNA. Results are representative of five sets of experiments.