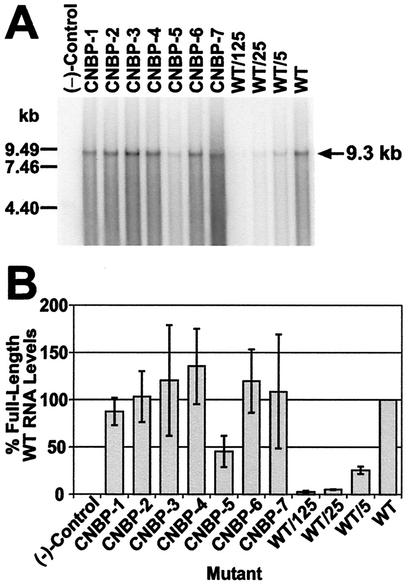
FIG. 2.
Hybridized RNA blot and phosphorimager analysis of NC mutant and wild-type HIV-1 transiently expressed in 293T cells. (A) Particles were analyzed for full-length HIV-1 genomic RNA by using a 32P-labeled probe comprised of an 8.1-kbp AvaI pNL4-3 fragment. All samples were adjusted for equal RT levels (4.1 × 106 cpm), fractionated, and treated as described in Materials and Methods. Dilutions (undiluted, 5-, 25-, and 125-fold) of the wild-type sample were also tested. (−)-Control, samples isolated and prepared from centrifuged supernatants from 293T cells transfected with sheared salmon sperm DNA. The RNA markers are indicated on the left and the size of full-length HIV-1 RNA genome is noted on the right. (B) Band intensities were obtained by phosphorimager analysis from Northern blots (performed as described in Materials and Methods and in part A of this figure) from three independent transfections. The samples for each Northern blot were adjusted for equal levels of RT activity. The intensity of the undiluted wild-type sample (WT) was set to 100% and the intensities of the remaining samples and dilutions of the wild-type sample were calculated relative to the undiluted wild-type sample. The percentage of full-length wild-type RNA levels obtained is representative of the average of three independent RNA blots. Error bars depicting the standard deviation from the mean are also presented. (−)-Control is as described for panel A.