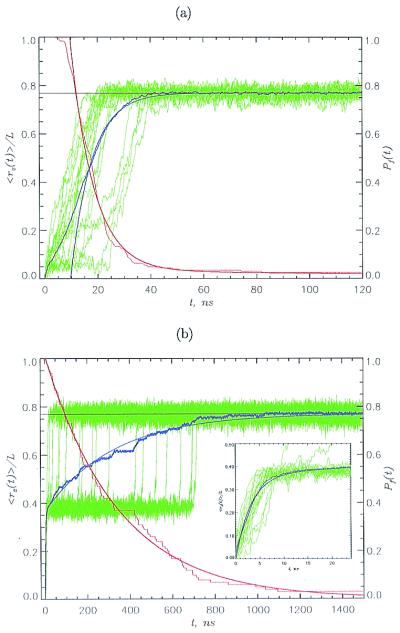
Figure 2.
Unfolding kinetics at constant force. We used fs = 69 pN for S1 (a) and S2 (b). The normalized z component of the end-to-end distance <rz(t)>/L, given in blue, is averaged over 50 trajectories. Smooth continuous blue curves are single (a) or double (b) exponential fits. Data in green show the extension for 20 individual trajectories. The fractions of folded molecules Pf(t) are given in red, and smooth red curves are double (a) and single (b) exponential fits. The unfolding times are given roughly by Poisson distribution. The large heterogeneity in unfolding times is surprising, because the initial state in unfolding simulations is a relatively well defined folded conformation. Horizontal black lines are the equilibrium extensions under simulation conditions. S1 unfolds by an unzipping motion in a single kinetic step in which all strands unfold simultaneously; S2 reaches S in two steps. An intermediate I [<rz(t)>/L ≈ 0.4] is quickly formed, and the transition I → S occurs on a longer time scale. I is an on-pathway intermediate that exists in all unfolding trajectories. The dynamics of N → I are shown (Inset).