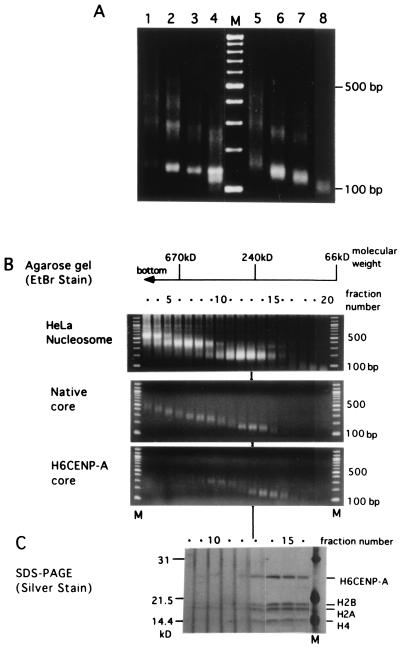
Figure 5.
MNase digestion of the reconstituted nucleosomes and glycerol gradient sedimentation of the MNase digests. (A) Nucleosomes were formed from native core histones (lanes 1–4) or His6CENP-A/core histones (lanes 5–8). Aliquots of 8 μl were digested with 0.01 unit (lanes 1 and 5), 0.03 unit (lanes 2 and 6), 0.1 unit (lanes 3 and 7), or 0.3 unit (lanes 4 and 8) of MNase for 10 min at 37°C. The DNA of each sample was electrophoresed through a 1.7% agarose gel and detected by ethidium bromide fluorescence. Lane M shows a 100-bp ladder. (B) Glycerol density gradient sedimentation of the MNase digests of the nucleosomes from HeLa nuclei (Top), reconstituted with native core histones (Middle), or His6CENP-A core histones (Bottom). Each 150-μl aliquot was fractionated from the bottom, and 20 μl of each fraction was electrophoresed through 1.7% agarose gel after proteinase K digestion. Each lane of B and C was numbered according to the fraction number of the glycerol gradient. The position of each molecular mass marker (albumin, 66 kDa; catalase, 240 kDa; thyroglobulin, 670 kDa) was marked at the top. M, 100-bp ladder. (C) The remaining 130 μl of glycerol gradient fractions 8–16 (B Bottom) were precipitated with acetone and separated by 15% SDS/PAGE. The protein bands were detected with silver staining.