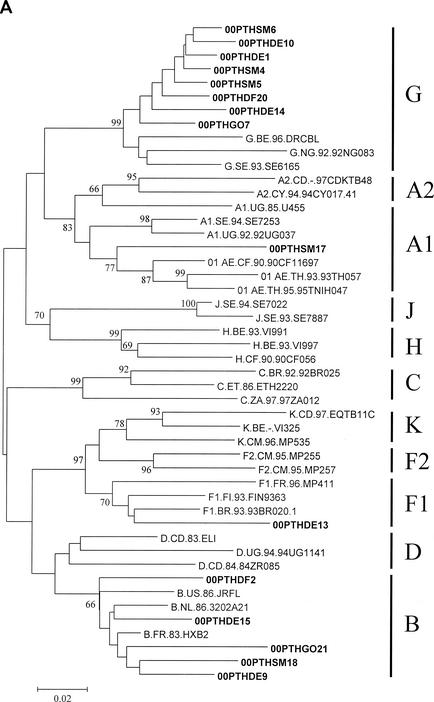
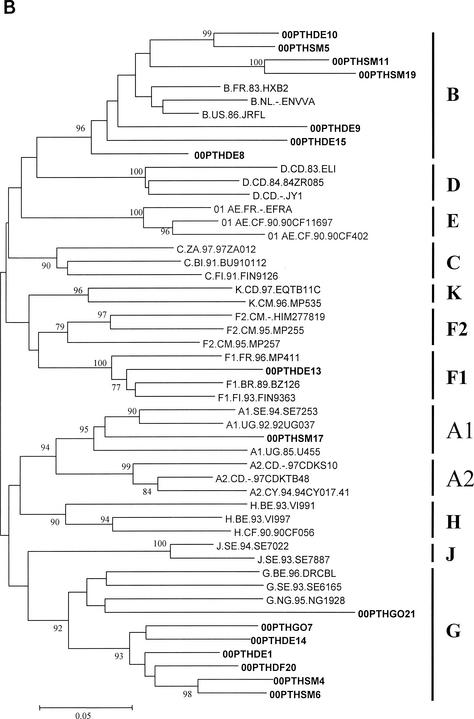
FIG. 1.
Genetic subtypes and evolutionary relationships of the virus sequenced in this study based on neighbor-joining phylogenetic trees of partial gag (A) sequences (p17) and partial env (B) sequences (C2-V3). The phylogenetic trees were constructed with reference sequences from all HIV-1 subtypes and subsubtypes as well as with the Portuguese (PT) sequences (shown in boldface type). The bootstrap values supporting each of the internal branches defining a subtype or subsubtype are shown. Bootstrap values of 70% or greater provide reasonable confidence for assignment of an individual segment to one or the other genotype. The scale represents genetic distance.