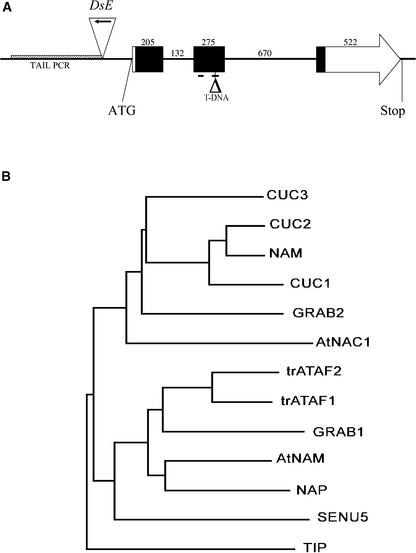
Figure 2.
The CUC3 Gene and Predicted Protein.
(A) Structure of CUC3. Boxes represent the open reading frame, the arrow at the end of the box at right corresponds to the C terminus of the predicted CUC3 protein, and lines indicate introns or untranslated regions. Black boxes represent nucleotides that encode the NAC domain, and numbers represent the lengths of exons and introns (in nucleotides). Lines below the second exon indicate putative bipartite nuclear localization signals. ATG and Stop indicate the start and stop codons; the triangle at left (with arrow) represents the DsE element in the WET368 (cuc3-1) line, and the arrow indicates the GUS gene; the shaded box represents the thermal asymmetric interlaced (TAIL) PCR product used to isolate CUC3; and the triangle at right indicates the Versailles T-DNA insertion in cuc3-2.
(B) Phylogenetic tree of the NAC domains of Arabidopsis CUC3, CUC2, CUC1, AtNAC1, ATAF1, ATAF2, AtNAM, NAP, and TIP, petunia NAM, wheat GRAB1 and GRAB2, and tomato SENU5. ATAF1 and ATAF2 are preceded by “tr” to indicate that the protein sequences were obtained by translation of mRNA sequences, leading to predicted proteins that are 60 amino acids longer than the protein sequences stored in GenBank.