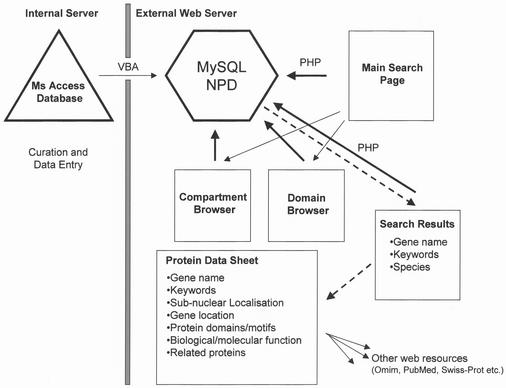
Figure 1.
Overview of the Nuclear Protein Database (NPD). Data is entered into a dedicated MS Access database (triangle) and once a month the Access data tables are converted to MySQL tables using MS Visual Basic (VBA). The SQL tables are then copied to the external MySQL server (hexagon) to be queried using PHP (thick arrows). NPD can be searched using Boolean operators via the web using the main search page. Alternately, the database may be queried by compartment (Compartment Browser) or protein domain (Domain Browser). Search results (dashed arrows) are returned as a meta-list of matching gene entries. The individual entries are retrieved by clicking on the main gene name, which queries the MySQL server once again returning a protein data sheet (PDS) containing archived and external database information. All result and PDS web pages are generated dynamically using PHP. Web pages are represented by squares.