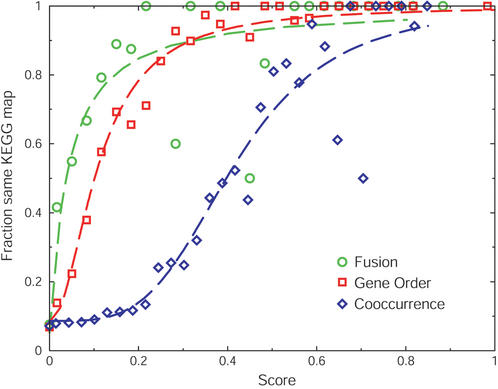
Figure 2.
Comparing different types of genomic associations to obtain equivalency. Scores are plotted versus the observed accuracy for each genomic association method. Data points indicate the fraction of predicted pairs of orthologous groups that are on the same KEGG map for each type of genomic association. For fusion and gene order, scores indicate the number of non-redundant observations divided by the number of species that contain at least one of the orthologous groups. The dashed lines in the respective colors represent fits of these data to standard saturating hill equations: f(x)=a+[(1−a)xb/(cb+xb)], where x represents the score, a the intercept, b the cooperativity, and c the value of x where half of the maximum is reached.