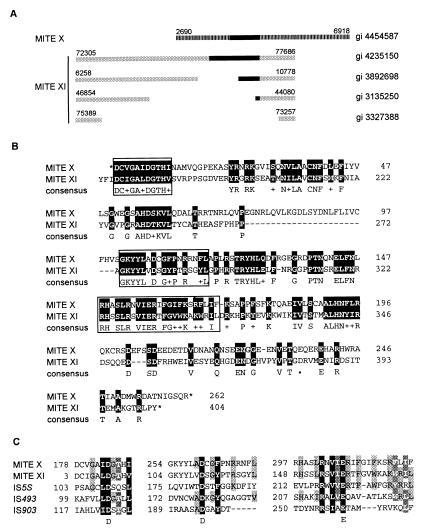
Figure 4.
MITE transposases (A). Diagram depicting structural similarities between members of MITE XI elements (gray boxes) and a member of MITE X (striped box). Black boxes represent ORFs corresponding to a putative transposase: MITE XI ORFs share >98% amino acid sequence similarity. MITE X and XI are distinct groups because no significant internal nucleotide sequence similarity is observed between MITE X and XI nor for the consensus sequence of their TIRs (5′-GG(G/T)GGTGTTATTGGTT-3′ for MITE X and 5′-GGCCCTGTTTTGTTTG-3′ for MITE XI). However, putative transposase ORFs, length of TIRs (16 bp), and TSD (5′-TTA-3′) of MITE X and XI are similar, indicating a related mechanism of transposition and possibly a common origin for both groups. GenBank gi numbers of the clones from which elements were mined are indicated to the right. Nucleotide positions of transposon termini are indicated above each element. (B) Amino acid sequence similarity between the conceptual translation for MITE X (gi 4454587, nucleotide position 4650–5865) and the corresponding region of MITE XI ORF (gi 4585884). Identical amino acids and functionally or structurally related residues are shaded in black. *, Translational stops. Boxed sequences indicate the region containing the DDE motif. (C) Alignment of conserved regions corresponding to the functionally important DDE motif found in transposases and integrases of many transposable elements (39–42). Transposases are from MITE X (gi 4454587, conceptual translation of nucleotides 4650–5865), MITE XI (gi 4585884), IS5S (gi 1256580) from Synechocystis sp., IS493 (gi 1196467), and IS903 (gi 136129) from Escherichia coli. Amino acid residues are shaded based on the level of structural and functional similarities. Residues conserved between all, four, or three sequences are shaded black, dark gray, and light gray, respectively.