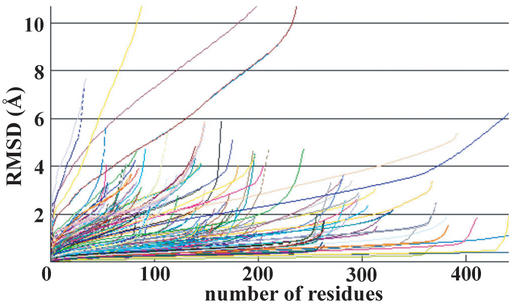
Figure 5.
Hubbard plots of 212 modeled 3D structures and real structures with sequence identity of more than 25%. The 3D structures of 212 proteins were determined after building enlarged FAMSBASE. The horizontal axis is the number of superimposed residues and the vertical axis is the best root mean square deviation given by the number of superimposed residues. A precise 3D model has a small RMSD for superimposition of many residues. An unreliable 3D model has a large RMSD for superimposition of a few residues. See reference 20 for detail.