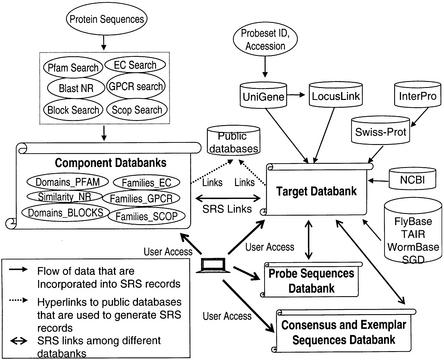
Figure 1.
Netaffx data flow. Protein sequences are annotated via the GRAPA battery of HMMs and PSI-BLAST models, in addition to Pfam, BLAST and BLOCKS searches. These annotations are consolidated into a unified XML format which is then indexed and loaded into SRS. These separate databanks (Domains_PFAM, Similarity_NR, Domains_BLOCKS, Families_EC, Families_GPCR, Families_SCOP) are summarized in the main Netaffx databank, the Target databank. Other annotations in the Target databank include those extracted from public databases according to the GenBank accession number of the representative sequences, as described in the text. Links exist from the Target databank to databanks of probe sequences and consensus and exemplar sequences. SRS databanks are indicated by the scroll-like icon.