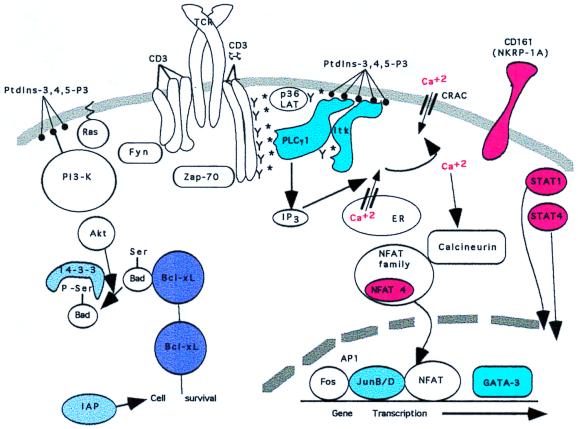
Figure 4.
A model for identified transcripts whose discordant expression is in accord with observed cell phenotypes. Pictured in cartoon format are genes whose transcripts have clearly defined cellular roles and whose discordant expression between GW4 and ME10 correlated with the established phenotypic differences. The genes are related by either being downstream of PI3-kinase, particularly several genes that regulate cell survival, or are directly required for calcium flux and calcium-regulated gene transcription. The proteins shaded various hues of blue represent genes expressed at significantly greater levels in GW4 (IL-4+) when compared with ME10 (IL-4-null). Conversely, those colored red were overexpressed in ME10 relative to GW4. Average copies of mRNA molecules per million for genes that were significantly altered by anti-CD3 treatment for GW4 (resting, activated) and ME10 (resting, activated), respectively, were: Itk (49, 115) and (25, 19); GATA3 (13, 26) and (8, 11); Jun-B (15, 118) and (14, 30); Jun-D (107, 291) and (68, 130); NFAT4 (33, 35) and (64, 26); STAT4 (36, 29) and (76, 36); 14–3-3 (24, 45) and (83, 41); Bcl-XL (11, 50) and (6, 7); and IAP (14, 211) and (10, 41). Selected genes whose expression was constitutive but discordantly expressed for GW4 vs. MW10 (GW4, ME10) were NKR-P1A (29, 459) and STAT1 (27, 79).