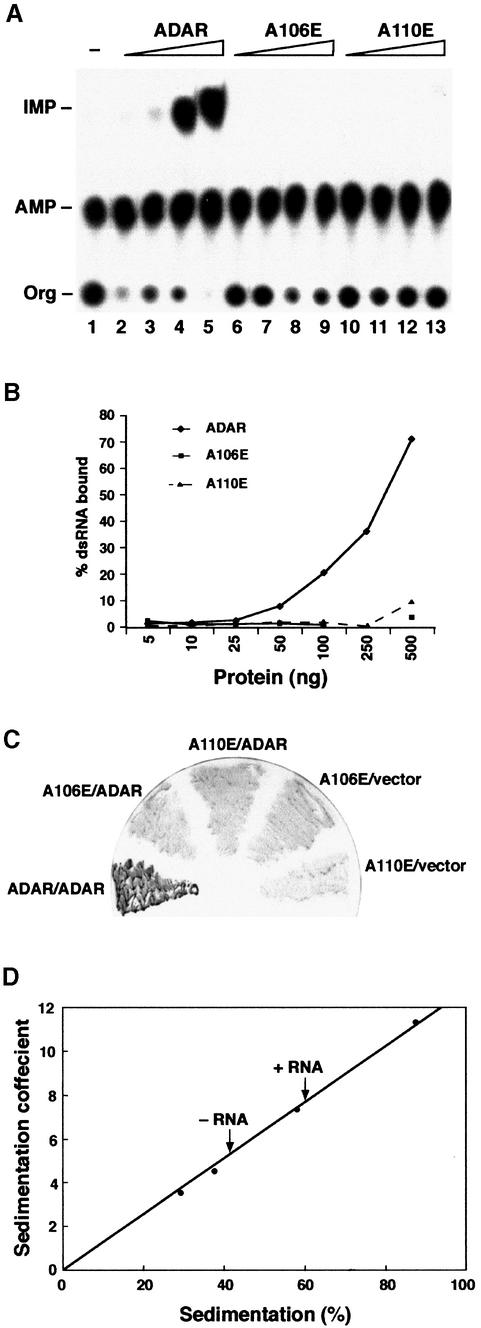
Fig. 4. dsRNA is required for ADAR dimerization. Single point mutations in dsRBD1 were generated so that amino acids A106 and A110 were changed to glutamate. (A) An in vitro editing assay was performed with recombinant proteins and chromatographed on a TLC plate. The positions of the origin (Org), adenosine (AMP) and inosine (IMP) are indicated. Increasing amounts of purified proteins were assayed [ADAR 3a (lanes 2–5), the mutant A106E (lanes 6–9) and the mutant A110E (lanes 10–13)], and the protein amounts were 10, 23, 70 and 140 ng, respectively. dsRNA without protein is in lane 1. (B) Filter binding was performed with the same mutant proteins A106E and A110E as well as with ADAR 3a; the protein amounts are indicated on the x-axis in nanograms. (C) Two-hybrid interactions between ADAR-LexA and ADAR-AD (positive control) and the two point mutants expressed as LexA fusion proteins and ADAR-AD were detected as blue β-galactosidase activity by filter lift assay. (D) Sedimentation coefficient of Drosophila ADAR 3/4-E/A in the presence or absence of RNA. Protein standards were applied to parallel gradients, and dADAR was detected by western blot analysis. The position of each protein is expressed as a percentage of the total number of fractions recovered from the gradient. The position of dADAR with and without RNA is indicated by arrows.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.