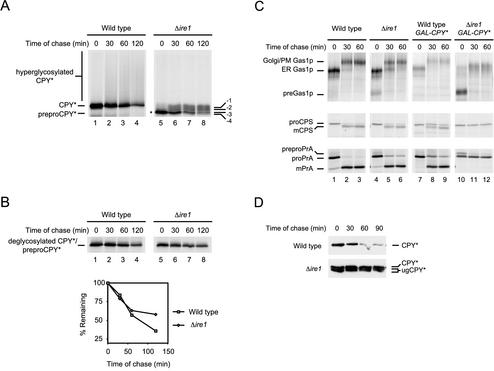
Figure 3.
CPY* overexpression causes the loss of multiple functions in Δire1 cells. (A) Wild-type and Δire1 cells overexpressing CPY* were pulse-labeled for 10 min with [35S]methionine/cysteine followed by a cold chase as indicated. CPY* was immunoprecipitated from detergent lysates and resolved by SDS-PAGE. The positions of untranslocated CPY* (preproCPY* and asterisk), underglycosylated CPY* (-1, -2, -3, -4), CPY*, and hyperglycoslyated CPY* are indicated. (B) CPY* was deglycosylated using Endo H but otherwise prepared and analyzed as described in A. Quantification of CPY* decay was performed by phosphorimager analysis (bottom panel). (C) Wild-type and Δire1 cells nonexpressing or overexpressing CPY* were pulse-labeled for 10 min with [35S]methionine/cysteine followed by a cold chase at the indicated times. Endogenous Gas1p, PrA, and CPS were immunoprecipitated from detergent lysates, separated by SDS-PAGE, and visualized by autoradiography. For CPS and PrA, carbohydrates were removed using Endo H to unambiguously identify the relevant species. The underglycosylation defect of Δire1 cells increased the complexity of some forms. Positions of untranslocated (pre-), ER, Golgi/plasma membrane (Golgi/PM), and mature forms (mCPS and mPrA) of each protein are indicated. (D) CPY* stability was monitored using cycloheximide chase analysis. Equal cell numbers of wild-type or Δire1 strains overexpressing CPY* were collected at the indicated times after the addition of cycloheximide. Proteins from cell lysates were separated by SDS-PAGE and analyzed by immunoblotting. Positions of CPY* and underglycosylated CPY* are indicated (CPY* and ugCPY*).