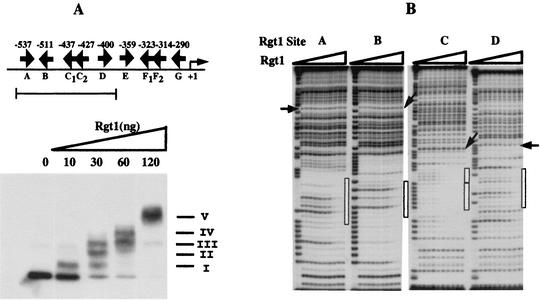
FIG. 3.
Binding of Rgt1 to the HXT3 promoter. (A) EMSA was carried out with a DNA fragment containing five Rgt1 binding sites. A constant amount of 32P-labeled DNA fragment (3 × 104 cpm) was incubated with increasing amounts of Rgt1 as indicated above the gel lanes. Arrows, locations and orientations of Rgt1 binding sites. Roman numerals denote Rgt1-DNA complexes. (B) The five Rgt1 sites were separated and used as probes for DNase I footprinting assays. After incubation of a 32P-labeled DNA fragment (2 × 105 cpm) with increasing amounts of Rgt1 (1, 3, 6, 10, 30, 60, 100, 300, and 600 ng per lane, from the third gel lane from the left) for 20 min, the DNA-protein complexes were digested with 2 U of DNase I for 1 min. The first and second gel lanes are the A+G ladder and the control (without Rgt1), respectively. Site C consists of two Rgt1 binding sites, C1 and C2, 3 bp apart. The binding affinities of Rgt1 were estimated by comparing the intensities (measured on a PhosphorImager by the ImageQuaNT program) of a reference band (indicated by arrows) and a band within the Rgt1 footprint (indicated by boxes).