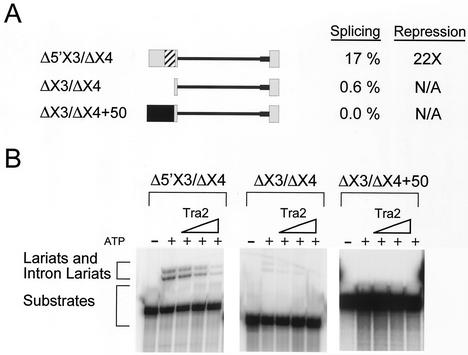
FIG. 5.
Identification of sequences in exon 3 required for M1 splicing. (A) RNAs used for in vitro splicing assays. Boxes, exons; lines, M1 intron; thick line, substituted 3′ splice site region from ftz RNA; striped box in exon 3, position of the ACE. Δ5′X3/ΔX4 includes 58 nt from the promoter-distal end of exon 3, the M1 intron, and the first 24 nt of exon 4, while ΔX3/ΔX4 contains only the last 8 nt of exon 3, M1, and the first 24 nt of exon 4. ΔX3/ΔX4+50 contains an additional 50 nt of plasmid sequences (black box). The in vitro splicing efficiencies as a percentage of products and intermediates and the amounts of repression observed when maximum Tra2 protein is added (3 pmol) are indicated to the right. (B) Lariat products and intermediates from in vitro splicing reactions performed on the constructs in panel A in the presence of increasing amounts of Tra2 (0.5, 2.5, and 12.5 pmol). Control reactions without added Tra2 both with (+) and without ATP (−) are also shown for each substrate.