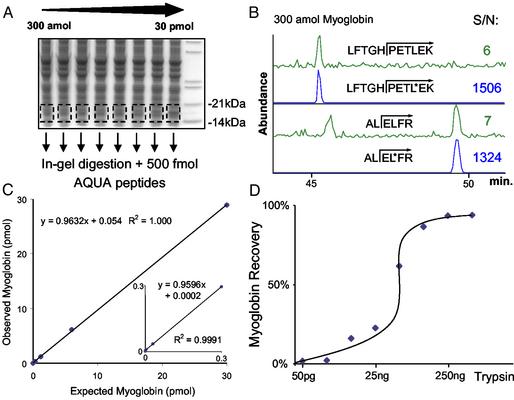
Fig. 3.
Validation of the AQUA method for horse heart myoglobin. (A) SDS/PAGE gel separation of 50 μg of yeast lysate spiked with standardized myoglobin protein at different amounts (300 amol to 30 pmol). Regions corresponding to migrated myoglobin were generously excised from the gel and digested with trypsin in the presence of 500 fmol of each AQUA internal standard peptide. (B) Analysis of 300 amol of myoglobin from a yeast background using the two AQUA peptides for reference and quantification by LC-SRM. The top trace of each pair represents the response for the “native” peptide formed by trypsinization, and the bottom trace corresponds to the same peptide synthesized with stable isotopes to have a mass difference of 7 Da. The peak area signal-to-noise is indicated for each determination. (C) Observed vs. expected response curve for the myoglobin quantification. (Inset) The low end of the curve (300 amol–300 fmol). (D) Effect of trypsin amount (50 pg to 1 μg) on the levels of myoglobin detected from the SDS/PAGE gel during a 6-h digestion. The same experiment as in A was performed, except the amount of spiked myoglobin (300 fmol) and AQUA peptides (500 fmol) was held constant while the amount of trypsin added for in-gel digestion varied. The curve plateaus when >250 ng trypsin was added, suggesting complete trypsinization.