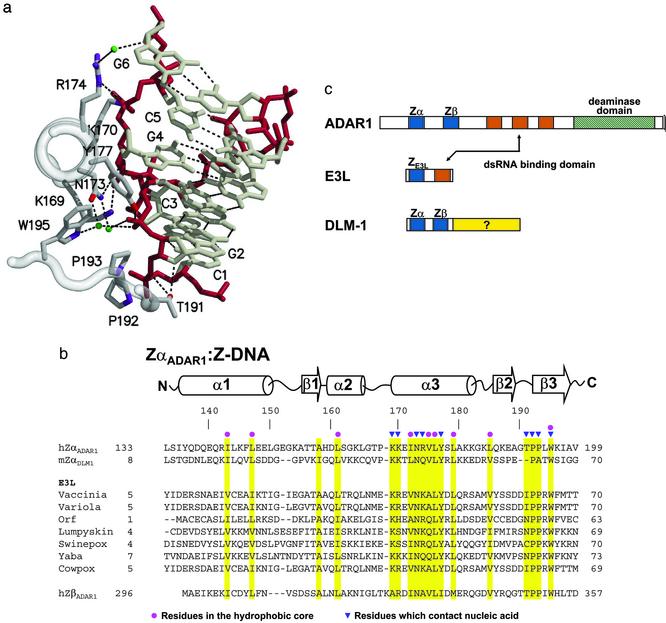
Fig. 1.
A family of Z-DNA-binding proteins. (a) DNA–protein contacts. A portion of the Z-DNA-binding domain of the ADAR1 editing enzyme (ZαADAR1) is shown complexed to a fragment of Z-DNA, as revealed in the structure of the cocrystal (10). The view is down the recognition helix of the protein (left), and a number of amino acids are shown that interact with left-handed Z-DNA stabilized by electrostatic and van der Waals interactions. The Z-DNA backbone is red and water molecules are green. There is an edge-to-face van der Waals interaction between guanine 4 and tyrosine 177. Note the van der Waals interactions between prolines 192, 193, and the Z-DNA backbone. (b) The amino acid sequences of Z-DNA binding and related proteins are shown underneath the secondary structure diagram, as revealed in the cocrystal structures of human ZαADAR1 (10) and mouse ZαDLM1 (11). A number of poxvirus E3L sequences are listed, as is the related sequence of human ZβADAR1. Yellow bars indicate residues important for the protein fold (red dots) and for Z-DNA recognition (blue triangles). The amino acid numbering at the beginning and the end of each sequence is indicated. The GenBank accession numbers for the various sequences are as follows: double-stranded RNA adenosine deaminase, AAB06697.1 (Homo sapiens); Z-DNA-binding protein 1; tumor stroma and activated macrophage protein DLM-1 (Mus musculus) NP_067369; the E3L proteins, AAA02759 (vaccinia virus); NP_042088 (variola virus); AAC08018 (orf virus); AAK84995 (lumpy skin disease virus); NP_570192 (Swinepox); NP_073419 (yaba-like disease virus); and CAC42100 (cowpox virus). (c) Three different families of proteins are illustrated, with domains shown as colored boxes. The Zα and Zβ domains of both ADAR1 and DLM1 are blue, as is the related domain of E3L. Vaccinia E3L protein and the editing enzyme ADAR1 both have domains that bind to double-stranded RNA, shown in red.