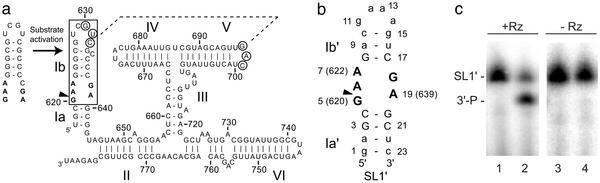
Fig. 1.
(a) Sequence and secondary structure of the Neurospora VS ribozyme. The interaction between stem-loops I and V is indicated by a dashed line, and residues involved in this interaction are circled (7). Upon tertiary folding of the ribozyme, stem-loop I (subdivided into Ia and Ib) undergoes a structural change from an inactive to an active conformation (11). The minimal substrate domain for the trans cleavage reaction is boxed (10), and the cleavage site is indicated by the arrowhead. (b) Sequence and secondary structure of SL1′ RNA. WT and mutant nucleotides are represented by uppercase and lowercase letters, respectively. (c) SL1′ is cleaved by the VS ribozyme. 3′-32P-end-labeled SL1′ (25 nM) was incubated at 30°C in 40 mM Tris·HCl (pH 8.0), 50 mM KCl, and 100 mM MgCl2 in the presence (lanes 1 and 2) or absence (lanes 3 and 4) of unlabeled RZ6 ribozyme (51) (37 μM) for 0 h (lanes 1 and 3) or 16 h (lanes 2 and 4) (12). Full-length SL1′ and the 3′ product (3′-P) were separated by denaturing PAGE and visualized with a PhosphorImager.