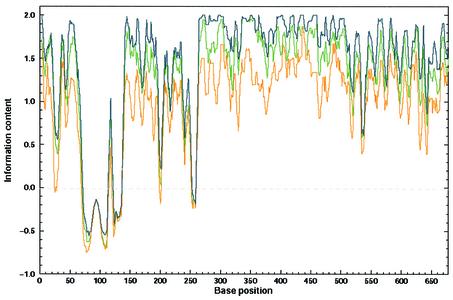
Figure 1.
DNA conservation of nef DNA. To compare the information content of nef DNA sequences with encoded Nef proteins, we used a back-translation technique. The correct back-translation was made by replacing every amino acid by the corresponding codon from the real, known gene sequence; the randomized back-translation was made by replacing every amino acid by one of the corresponding randomly selected codons of its own repertoire (equiprobable within the group). The codon usage-related back-translation was made by using EMBOSS Backtranseq according to the HIV-1 codon usage file: EHuman_immunodeficiency_virus_type_1.cut. The curves showing the information content were smoothed by a running average with a window size equal to six. The green line represents the DNA conservation of the nef sequences retrieved from the original database. The blue line describes the conservation of the codon usage-related back-translated nef sequences, and the orange line describes the conservation of the random back-translation. Conservation of DNA at every position was computed according to Equation 1.