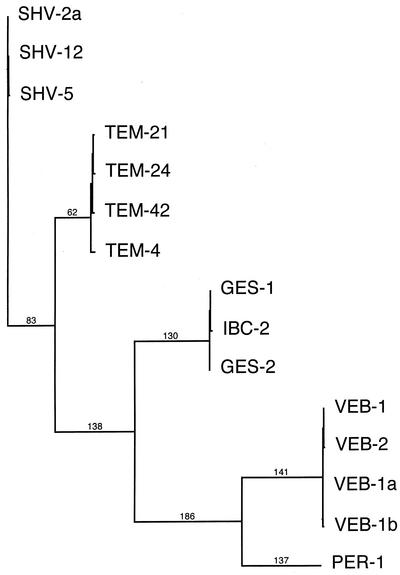
FIG. 1.
Dendrogram obtained for class A ESBLs identified in P. aeruginosa by parsimony analysis. The alignments used for construction of the tree were carried out with the ClustalW program, followed by minor adjustments to fit the class A β-lactamase scheme (1). Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The number of changes is indicated above each branch. The distance along the vertical axis has no significance.