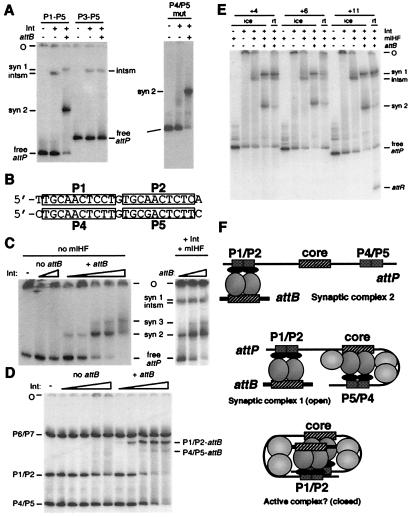
Figure 2.
The role of arm-type sites. (A) Formation of integration complexes with mutant attP substrates. Reactions were performed on ice as in Fig. 1B with an attP DNA containing P1–P5 or P3–P5 (Left) or mutant P4/P5 sites (Right), Int-L5 (as indicated), mIHF, and attB, as indicated. (B) Comparison of the sequences of the P1/P2 and P4/P5 arm-type sites. (C) Detection of a second mIHF-independent SC. SCs were formed on ice as in Fig. 1B by using attP, varying amounts of Int-L5 (Left; from left to right: 0, 0.24, 0.72, 0.024, 0.072, 0.24, 0.72, and 2.4 pmol Int-L5; these lanes contain 0.12 pmol attB per reaction) and varying amounts of attB (Right; from left to right: 0.06, 0.12, and 0.6 pmol attB per reaction; these lanes contain 0.24 pmol Int-L5 per reaction), in the presence or absence of mIHF as indicated. (D) P1/P2 sites prefer to bridge with attB. Radiolabeled DNA fragments containing only one pair of arm-type sites (P1/P2, 497 bp; P4/P5, 318 bp; P6/P7, 876 bp) and no core sites were mixed and incubated on ice in the presence or absence of 0.12 pmol of attB DNA (as indicated) and varying amounts of Int-L5 (−, no Int-L5 added; left to right: 0.024, 0.072, 0.24, 0.72, and 2.4 pmol Int-L5 per reaction) in the absence of mIHF under conditions identical to complex formation reactions. The positions of the P1/P2-attB complex, the P4/P5-attB complex, and the free P1/P2, P4/P5, and P6/P7 DNA fragments are indicated. (E) Effect of P1/P2-core intersite spacing on recombination and complex formation. Reactions were performed either on ice or at room temperature (as indicated) by using attP DNA fragments ranging from 451 bp to 458 bp, depending on the size of the insertion. Results of reactions with attP insertions of +4 bp, +6 bp, and +11 bp are shown, and reactions with insertions of +13 bp, +15 bp, +17 bp, and +21 bp were performed but are not shown. attP DNAs with insertions of +13 bp, +15 bp, and +17 bp did not recombine; however, those with insertions of +21 bp did, and none interfered with complex formation (data not shown). (F) Proposed structures of protein–DNA complexes. SC2 is postulated to contain Int-mediated bridges between attB and the P1/P2 arm-type sites. SC1 contains these same bridges but also contains Int-mediated, mIHF-stabilized intramolecular bridges between the attP core and the P4/P5 sites. Because the spacing changes shown in E do not affect complex 1 formation, we suggest that it is unfolded or open, such that the helical phasing of the P1/P2 sites and the core is not important. However, because nonintegral DNA insertions inhibit recombination, we propose that it is necessary for SC1 to fold into a more compact or closed structure in order for strand exchange to occur. The mIHF host factor is shown as light gray balls situated between the attP core and P4 in complex 1 and also between P2 and the core in the putative active complex. DNase I footprinting suggests that there may also be a unit of mIHF bound just to the left of the core in SC1 (7).