Table 1. Data collection, phasing, and refinement statistics.
| Remote | Edge | Peak | |
|---|---|---|---|
| Data collection and phasing | |||
| Wavelength, Å | 1.2565 | 1.2832 | 1.2825 |
| Resolution, Å | 2.0 | 2.0 | 2.0 |
| No. of total reflections | 497,657 | 364,430 | 299,731 |
| No. of unique reflections* | 97,852 | 97,565 | 96,091 |
| Completeness, % | |||
| Overall | 100.0 | 99.9 | 98.5 |
| Outer 0.1-Å shell | 100.0 | 99.9 | 93.1 |
| Rmerge† | |||
| Overall | 0.059 | 0.060 | 0.055 |
| Outer 0.1-Å shell | 0.329 | 0.342 | 0.269 |
| Mean figure of merit for MAD phasing‡ | 0.633 | ||
| Refinement statistics | |||
| No. of reflections, working set/testing set | 92,811/4,815 | ||
| Data cutoff, σ | 0.0 | ||
| R/Rfree§ | 0.199/0.211 | ||
| No. of protein atoms | 4,316 | ||
| No. of zinc ions | 7 | ||
| No. of ligand atoms | 32 | ||
| No. of water molecules | 309 | ||
| rms deviations | |||
| Bond lengths, Å | 0.006 | ||
| Bond angles, ° | 1.3 | ||
| Proper dihedral angles, ° | 23.5 | ||
| Improper dihedral angles, ° | 0.7 |
Friedel mates are treated as unique reflections for data reduction in MAD experiments
Rmerge = 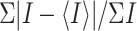 ,
where I is the observed intensity and
,
where I is the observed intensity and 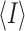 is the
average intensity calculated for replicate data
is the
average intensity calculated for replicate data
Mean figure of merit =
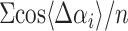 , where
Δαi is the error in the phase angle for
reflection i, and n is the number of reflections
, where
Δαi is the error in the phase angle for
reflection i, and n is the number of reflections
R =
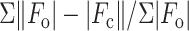 ,
where
,
where 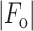 and
and 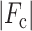 are
the observed and calculated structure factor amplitudes, respectively.
Rfree is calculated in the same manner for reflections in
the test set excluded from refinement
are
the observed and calculated structure factor amplitudes, respectively.
Rfree is calculated in the same manner for reflections in
the test set excluded from refinement
