Table 1. Crystallographic data, structure solution, and refinement statistics.
| DhaK-Dha complex | Ta6Br14 | KAu(CN)2 | PIP* | Apo-DhaK | |
|---|---|---|---|---|---|
| Data collection | |||||
| Resolution, ņ | 1.75 (1.78-1.75) | 2.45 (2.52-2.45) | 2.45 (2.55-2.45) | 2.45 (2.55-2.45) | 2.0 (2.04-2.00) |
| Unique reflections | 78,957 | 29,777 | 27,533 | 30,483 | 51,496 |
| Redundancy | 3.7 | 3.1 | 2.3 | 2.4 | 2.9 |
| Completeness, %† | 95.7 (97.5) | 99.8 (99.3) | 94.4 (90.4) | 96.2 (93.6) | 93.1 (60.6) |
| I/σ (|) | 19.4 | 21.3 | 23.4 | 21.6 | 23.7 |
| Rmerge, %†‡ | 4.6 (17.2) | 4.6 (7.4) | 3.5 (6.4) | 4.3 (7.9) | 4.6 (10.0) |
| Structure solution | |||||
| Resolution range, Å | 25.0-4.5 | 25.0-2.5 | 25.0-2.5 | ||
| Heavy-atom sites | 6 | 5 | 4 | ||
| Phasing power (acentric/centric) | 3.0/2.7 | 2.0/1.5 | 1.3/1.3 | ||
| RCullis (acentric/centric) | 58.1/55.4 | 67.1/69.1 | 81.7/79.6 | ||
| Refinement statistics | |||||
| Resolution range, Å | 25.00-1.75 | 25.00-2.00 | |||
| Rfactor§/Rfree,¶ % | 17.15/20.1 | 18.9/23.8 | |||
| rmsd bond lengths, Å | 0.012 | 0.018 | |||
| rmsd bond angles, ° | 1.618 | 1.780 | |||
| No. of reflections | 76,798 | 50,052 | |||
| No. of protein atoms | 5,026 | 5,026 | |||
| No. of water molecules | 508 | 485 |
rmsd, root-mean-square deviation from ideal geometry
PIP, Di-μ-iodobis(ethylenediamine) diplatinum (II)-nitrate
Numbers in parentheses refer to the indicated highest resolution shell
Rmerge =
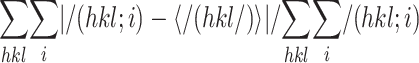 ,
where I(hkl; i) is the intensity of an individual
measurement and <I(hkl)> is the average intensity from
multiple observations
,
where I(hkl; i) is the intensity of an individual
measurement and <I(hkl)> is the average intensity from
multiple observations
Rfactor =
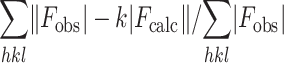
Rfree = the R factor against 5% of the data removed prior to refinement
