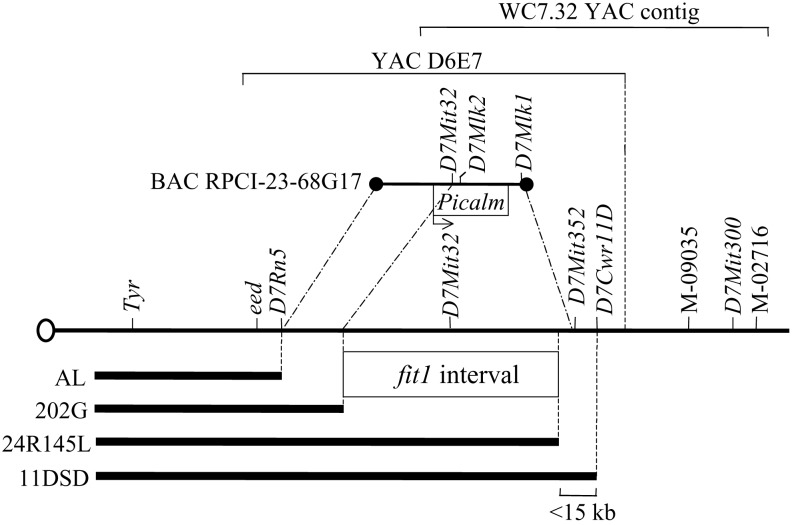
Fig. 1.
Genetic and physical map of a segment of mouse chromosome 7 containing the fit1 locus. Chromosome 7 is indicated by the line with the oval (depicting the centromere) at its left end. Relative locations of genetic loci (italicized) and STS markers (M-09035 and M-02716) are shown above the chromosome. No physical distance is implied by placement of any of the markers. Bold lines below the chromosome represent Del(Tyr) deletions, and their allele names (21) are shown (Left). The minimal deletion interval containing the fit1 locus (15) is indicated. A YAC contig spanning part of the region is shown above the chromosome [390-kb YAC clone D6E7 (18); WC7.32 contig is described at www.informatics.jax.org/searches/contig.cgi?1251 (660 kb from D7Mit32 to M-02716)]. D7Rn5 and D7Cwr11D are loci derived from the indicated deletion breakpoints. The RPCI-23-68G17 BAC clone (194 kb) containing the Picalm gene (with the transcriptional orientation shown) and several markers is shown above the chromosome. The lines projecting from the ends of the BAC to the chromosome indicate that the BAC is located between, but does not include, the D7Rn5 and D7Mit352 loci. The portion of the BAC between its distal end and the dashed/dotted line extending from the D7Mit32 locus indicates the minimum part of the BAC known to be located within the fit1 interval.