Fig. 1.
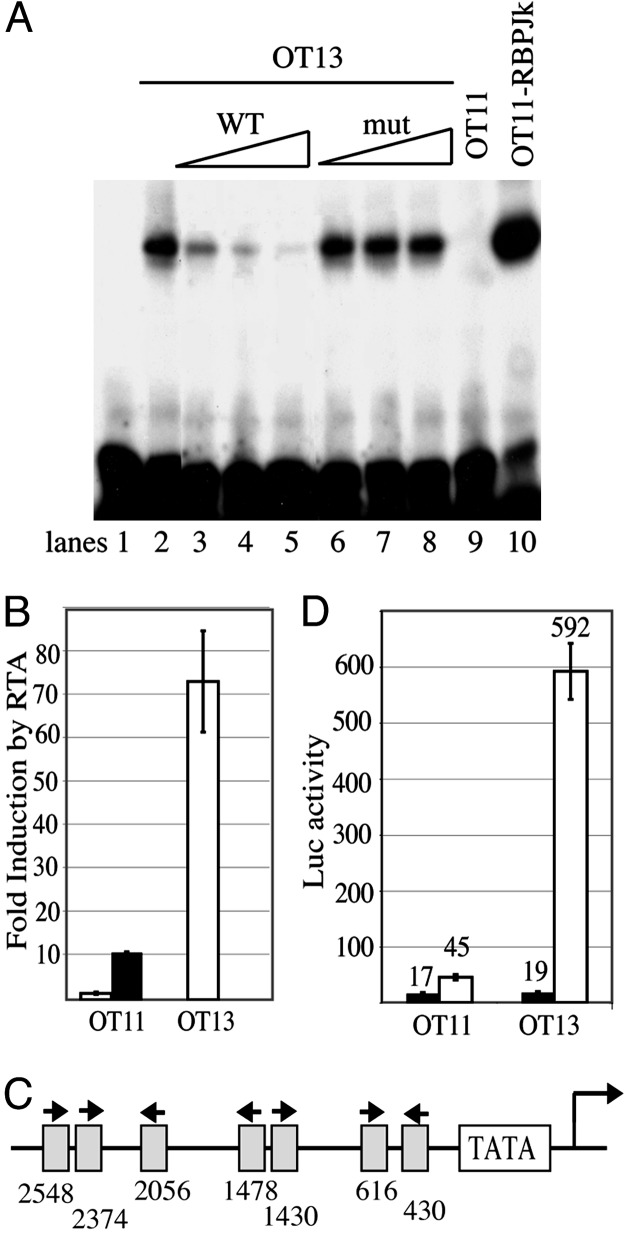
OT11 cells lack functional RBP-Jκ activity, and the RTA promoter contains functional RBP-Jκ sites. (A) Complexes able to specifically supershift RBP-Jκ oligos are present in OT13 but absent in OT11 nuclear extracts. Nuclear extracts were prepared as described in Materials and Methods. 32P-labeled RBP-Jκ oligos were incubated with nuclear extracts from OT13 (lanes 2–8), OT11 (lane 9), or OT11-RBPJκ (lane 10) in the absence (lane 2) or presence of increasing amounts of competitor oligos, WT (lanes 3–5) or mutant (mut) competitors (lanes 6–8). (B) RTA activation of RBP-Jκ-driven LUC reporter in OT11 cells is defective, and the defect can be partially corrected by cotransfecting an RBP-Jκ expression vector. Cells were cotransfected with template DNA 4×RBPJrev-Luc and effector DNA pcDNA3-RTA. In OT11 cells, RTA transactivation was tested in the absence (open bar) or presence (filled bar) of cotransfected pcDNA3.1-RBP-Jκ. LUC assay was performed as in Materials and Methods. The fold induction by RTA was plotted, with the error bars representing SDs of the results from at least two independent experiments. (C) Schematic depiction of putative RBP-Jκ-binding sites in the RTA promoter. The start of the RTA transcript is indicated by an arrow. The orientation and location of each RBP-Jκ-binding site upstream of the RTA transcript start site are shown. (D) RTA activation of RTA-promoter-driven LUC reporter was examined in either OT11 or OT13 cells. Cells were cotransfected with RTA-LUC and RTA (open bar) or empty vector (filled bar). pcDNA3.1-lacZ was included as an internal control. The LUC activities, normalized for transfection efficiency by β-galactosidase activities, are plotted, with the error bars representing SDs of the results from at least two independent experiments.