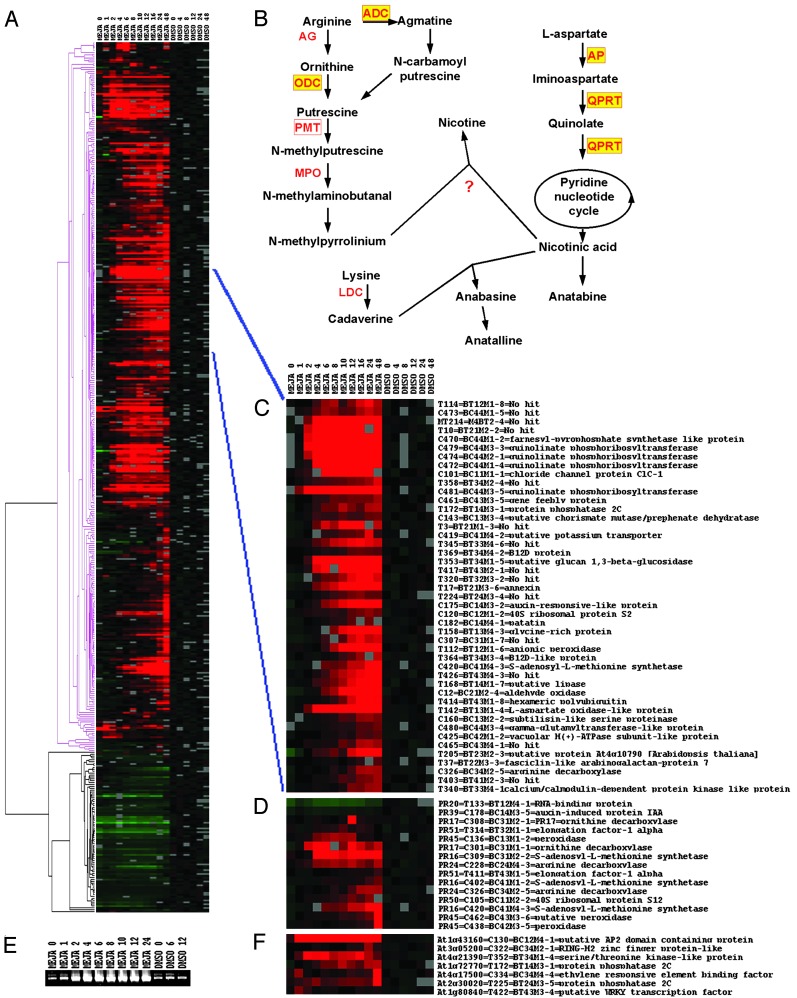
Fig. 2.
Transcriptome of jasmonate-elicited BY-2 cells. (A) Average linkage hierarchical clustering of MJM gene tags. The time points for the cluster analysis are indicated (in hours) at the top. Red and green boxes reflect transcriptional activation and repression, respectively, relative to the average expression level in control (DMSO) cells. Gray boxes correspond to missing time points. Pink and black parts of the dendrogram indicate the clusters of activated and repressed genes, respectively. (B) Biosynthetic pathway of nicotine alkaloids. Enzymes involved in the metabolic conversions are in red, those for which the corresponding tobacco gene or orthologs from other plants have been cloned are boxed, and those for which a corresponding MJM tag has been identified in the transcript profiling analysis are in yellow. (C) Subcluster of the MJM transcriptome. Shown is a close-up of a subcluster comprising gene tags coregulated with genes known to be involved in nicotine biosynthesis. (D) Average linkage hierarchical clustering of tags common between the MJM transcriptome and the genes differentially expressed in tobacco roots (50). The corresponding protein (PR) code, as used previously (50), has been included. (E) Expression analysis of pmt. Expression of pmt was verified by RT-PCR analysis on the RNA prepared for cDNA-AFLP transcript profiling and using oligonucleotides spanning nucleotides 8–897 of the N. tabacum cv. Burley pmt ORF (38). Time points are indicated (in hours) at the top of the ethidium bromide-stained gel. (F) Average linkage hierarchical clustering of signal transducing genes common between the MJM transcriptome and the wound-induced Arabidopsis transcriptome (33). The corresponding Munich Information Center for Protein Sequences code has been included. ADC, arginine decarboxylase; AG, arginase; AP, aspartate oxidase; LDC, lysine decarboxylase; MEJA, methyl jasmonate; MPO, methylputrescine oxidase; ODC, ornithine decarboxylase; PMT, putrescine methyltransferase; QPRT, quinolinate phosphoribosyltransferase.